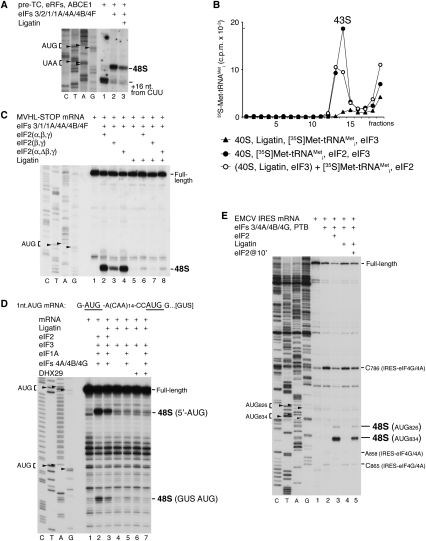
Figure 3.
Activity of Ligatin in 48S complex formation on mRNAs translated by the 5′-end-dependent scanning mechanism and on the EMCV IRES. (A) Toe-printing analysis of 48S complex formation on MVHL-STOP mRNA after incubation of pre-TCs, assembled on this mRNA, with eRFs, ABCE1, eIF2, eIF3, eIF1, eIF1A, eIF4A, eIF4B, and eIF4G in the presence/absence of Ligatin. Positions of initiation and stop codons and assembled ribosomal complexes are indicated. Lanes C, T, A, and G depict corresponding DNA sequences. (B) Influence of Ligatin on eIF2-mediated 43S complex formation, assayed by SDG centrifugation. (C–E) Toe-printing analysis of 48S complex formation on MVHL-STOP mRNA (C), GUS mRNA with an unstructured 5′-UTR containing an AUG triplet 1 nt from the 5′ end followed by 14 CAA repeats (D), and EMCV IRES in the presence of 40S subunits, Met-tRNAMeti, Ligatin, eIF2, and other factors (E), as indicated. Positions of initiation codons and assembled ribosomal complexes are indicated. Lanes C, T, A, and G depict corresponding DNA sequences.