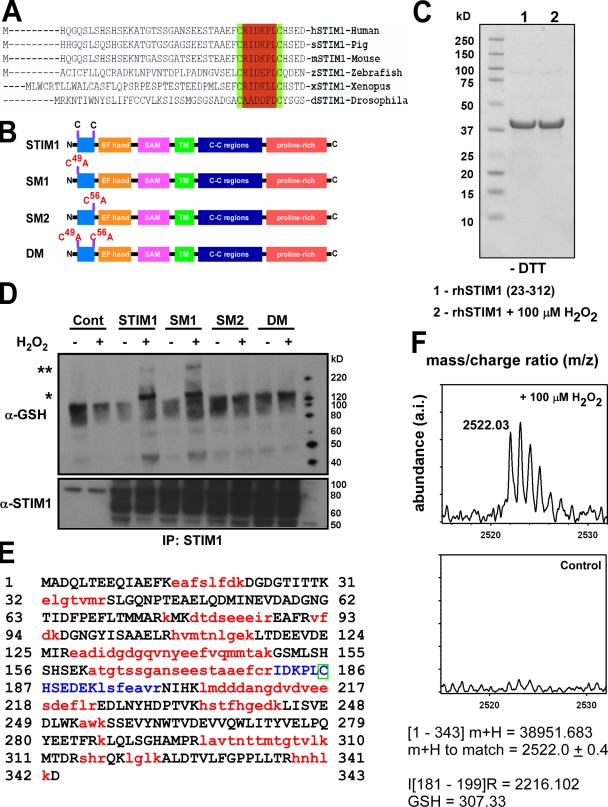
Figure 6.
Identification of cysteine 56 as the site for S-glutathionylation in response to oxidants. (A) Sequence alignment of STIM1 demonstrates evolutionary conservation of cysteine residues at positions 49 and 56. (B) Schematic for STIM1 mutant constructs. (C) Coomassie staining of a truncated N-terminal STIM1 fragment (amino acids 23–213) exposed to 100 mM H2O2 for 30 min and run on a 4–12% Bis-Tris gel under nonreducing conditions (−DTT). No shift in protein mobility was detected, indicating that oxidant stress did not facilitate the formation of disulfide bonds in the STIM1 protein. (D) COS7 cell lysates from STIM1-transfected cells were incubated with 200 µM H2O2 for 30 min and immunoprecipitated (IP) with an α-STIM1 antibody. Immunoprecipitated STIM1 was resolved by electrophoresis under nonreducing conditions (−DTT) and probed for S-glutathionylation using a small peptide antibody against GSH. Nontreated samples were also resolved under reducing conditions (+DTT) and probed for STIM1 for input control. A truncated N-terminal STIM1 fragment (amino acids 23–312) was subjected to tryptic digestion and assessed for S-glutathionylation at cysteine 56 by mass spectrometry. *, S-glutathionylation of STIM1; **, higher molecular mass STIM1 after H2O2 treatment. (E) Sequence of recombinant STIM1 fragment. Red indicates sequences not detected by MALDI time of flight. Blue indicates that the sequence contains cysteine 56. The green box indicates that the presence of GSH in this fragment can only be associated with the cysteine residue at position 56. (F) 30-min exposure of recombinant STIM1 protein to 100 µM H2O2 resulted in the formation of a mass spectra at a calculated mass of 2,522.0 ± 0.4 kD that was absent in the nontreated sample, corresponding to an increase of 306 to the predicted mass of the blue peptide fragment in D (2,216.102 kD). The molecular mass of reduced GSH is 307 D.