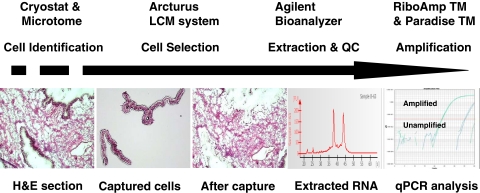
FIGURE 1.
The workflow of LCM. Tissue sections were cut using a cryostat or microtome and stained with hematoxylin and eosin (H&E) for cell identification. Pure epithelial cell populations from frozen ovarian tissue were captured using the Arcturus XT LCM system and visualized on the LCM cap. Total RNA was then extracted, and the quantity and quality of RNA were monitored on an Agilent bioanalyzer. Two peaks, 18S and 28S ribosomal RNA, were obtained in the RNA profile generated by the bioanalyzer, indicating successful RNA sample isolation. The RNA was amplified for downstream analysis. QC, quality control; qPCR, quantitative PCR.