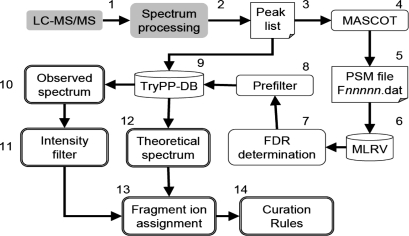
Fig. 2.
Workflow for automated annotation of phosphosites. Experimental LC–MS/MS data is gathered (1) and processed using platform specific software (2) to give a generic peak list file (3). This file, is used as the input to MASCOT (4), which generates a results file (5) containing all the PSMs. This file is parsed into the MLRV relational database (6) and the FDR for the search determined (7). A PSM-quality prefilter is applied (8) and suitable PSMs are exported to the TryPP-DB (9) where they are linked to the source peak list file used for the search. The observed MS/MS spectrum is extracted from the peak list file (10), filtered by an intensity threshold (11) and compared with a calculated fragmentation spectrum (12) for the peptide under examination. Observed ions are assigned to series (13) allowing the curation rules to be applied (14).