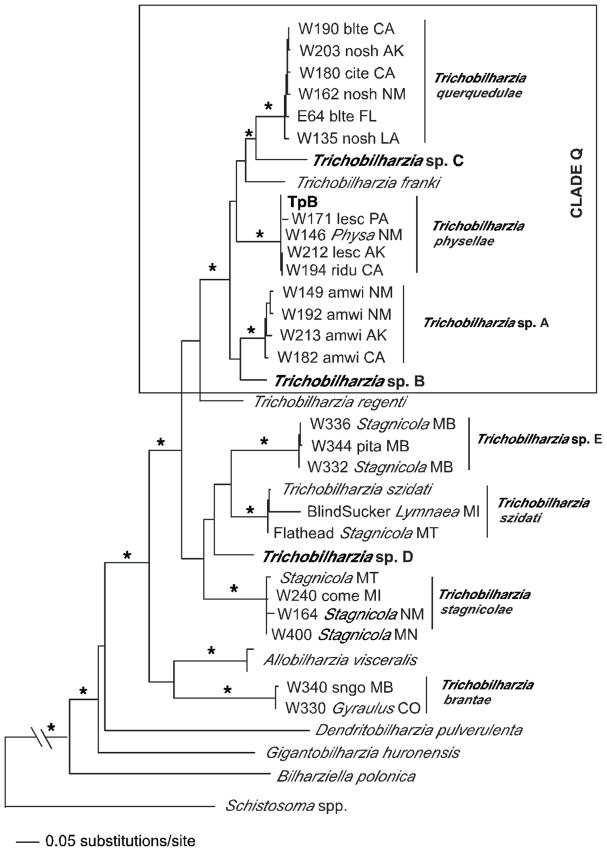
FIGURE 4.
Maximum likelihood tree based on cox1 sequences. For the Trichobilharzia querquedulae clade only some of the worms sequenced are represented as many differed by only one base pair. The “*” indicates node support of >95% bootstrap for MP and ME and >98 Bayesian PP. The ‘-’ indicates no significant node support. Outgroup species of Schistosoma were collapsed. For convenience, the following taxa were trimmed from the tree, but were fully supported in the clade: W137blteLA, W156blteNM, W148.1citeNM, W148.2citeNM, W155.3citeNM, W158noshNM, W162noshNM, W183noshCA, SDS1006noshNE, E45blteFL (Table III). The same was done for T. physellae, except in one case there were identical haplotypes: TpB = W171lescPA, W193lescNM, W255buheNM, W263PhysaNM. Otherwise, the following with only 1–2 bp differences were removed W211olsqAK, W193lescNM, W236PhysaMI, W230.1comeMI, and W256lescNM.