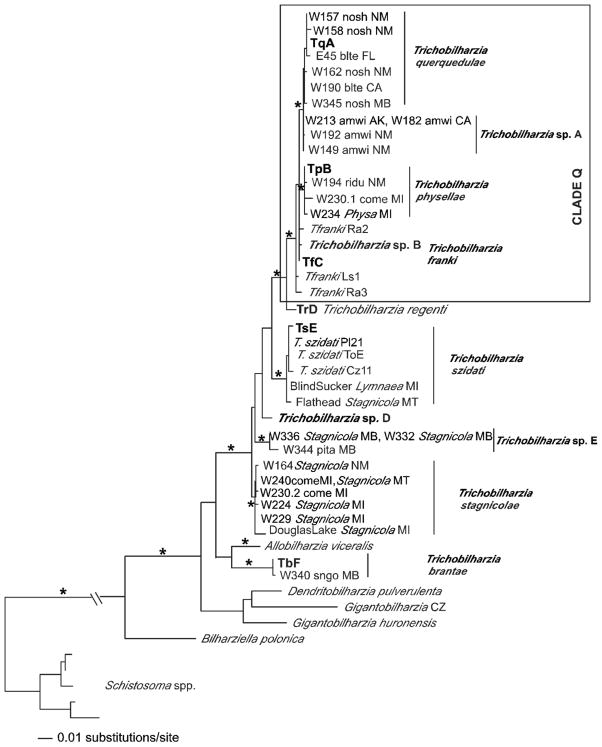
FIGURE 5.
Maximum likelihood tree based on ITS sequences. The following labels apply to samples with identical haplotypes: Trichobilharzia querquedulae TqA= W135blteLA, W137blteLA, W156blteNM, W148.1citeNM, W155.3citeNM, W180citeCA, W203noshAK, W183noshCA, SDS1006noshNE. Trichobilharzia physellae TpB = W146PhysaNM, W263PhysaNM, W171lescPA, W212lescAK, W249cabaNV, W255buheNM. Trichobilharzia franki TfC = Trichobilharzia sp. C, T. franki Ra1, and T. franki RSFO1. All haplotypes of T. regenti downloaded from GenBank were identical; TrD = T. regenti Cz79, T. regenti Cz31, T. regenti Pl27, T. regenti Pl20, T. regenti Pl17, T. regenti Pl14. Trichobilharzia szidati TsE = T. szidati Tsz, T. szidati Ls5, T. szidati ToA. Trichobilharzia brantae TbF = W346GyraulusMB, W331GyraulusCO, W330GyraulusCO. Isolates of T. franki are from R. ovata (ov) and R. auricularia (Ra) snails (one sample is from Lymnaea stagnalis = Ls). The “*” indicates node support of >95% bootstrap for MP and ME and >98 Bayesian PP. The ‘-’ indicates no significant node support.