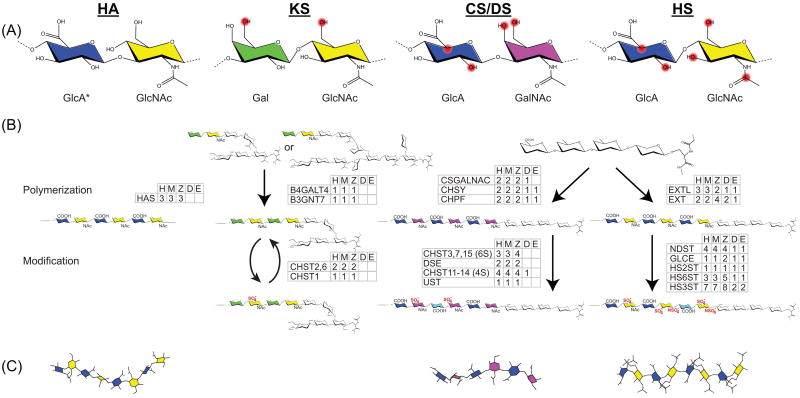
Figure 1. Interrelated biosynthesis of GAG classes yields similar yet distinct structures.
(A) GAGs are classified based on their disaccharide building blocks. Potentially modified positions are highlighted in red. (B) Numerous enzymes catalyze GAG elongation and modification. Families of enzymes involved in each step are listed with the number of homologs from humans-H compared to four models organisms: mouse-M, zebrafish-Z, Drosophila -D, and C elegans-E. Saccharides that link KS to core proteins are quite diverse and simplified here for clarity. Descriptions of each gene family along with specific gene IDs are in the Supplemental. (C) GAGs adopt distinct helical conformations. Illustrations were generated using Chem3D Pro with the pdb files 2BVK (HA), 2KQO (CS), and 1HPN (Heparin).
*Saccharides and modifications are abbreviated using the following nomenclature: GluA, β-D-glucuronic acid; GlcNAc, α/β-D-glucosamine; GalNAc, β-D-galactosamine; Gal, β-D-galactose IdoA, α-L-iduronic acid; NAc, N-acetylation; SO4−, sulfate. Specific disaccharides are condensed. For example, the disaccharide containing glucuronic acid linked to a 4-O and 6-O sulfated galactosamine is represented by GlcA-GalNAc4S, 6S.