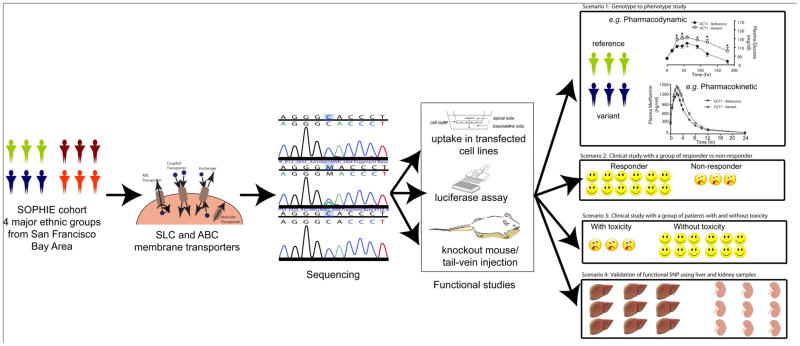
Figure 2.
An overview of the scientific approaches used by PMT in SNP discovery and functional studies. The PMT SNP discovery studies involve resequencing of coding and noncoding regions of membrane transporter genes in healthy volunteers in four major ethnic groups (the SOPHIE cohort). Various in vitro and in vivo assays are used to characterize coding and noncoding region variants in transporters. In order to validate functional findings and to translate them from bench to clinic, one or more of the following scenarios are applicable. (i) Hypothesis-driven clinical studies to evaluate the role of functionally important genetic variants in drug disposition and response in the SOPHIE cohort (figure from scenario 1 reprinted from ref. 8). (ii) and (iii) Collaborative clinical studies involving PGRN investigators, CALGB, and GAP-J, with a focus on the role of genetic variants in transporters in drug response and toxicity. (iv) Association of functional SNPs with expression levels of transporters in liver and kidney samples. ABC, ATP-binding cassette; CALGB, Cancer and Leukemia Group B; GAP-J, Global Alliance in Pharmacogenomics, Japan; PGRN, Pharmacogenetics Research Network; PMT, Pharmacogenomics of Membrane Transporters project; SLC, solute carrier superfamily; SNP, single-nucleotide polymorphism; SOPHIE, Study of Pharmacogenetics in Ethnically Diverse Populations.