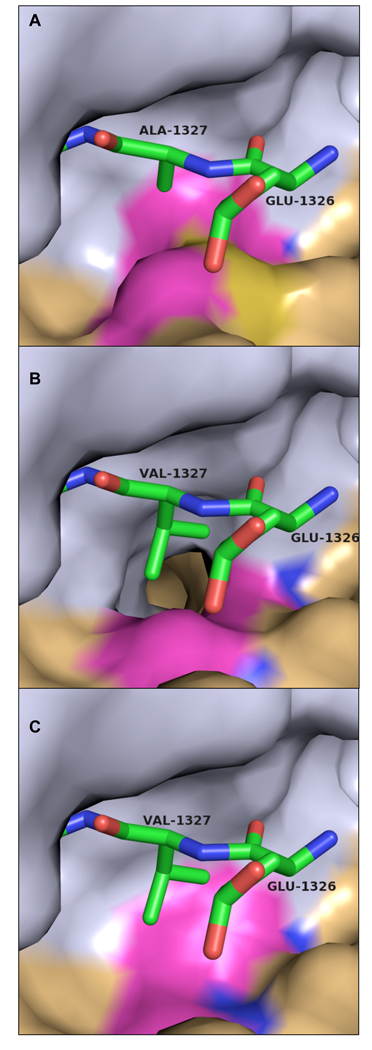
Figure 4. Covariance of amino acid substitutions in different alphaviruses strains at the S3 subsite and the P3 cleavage substrate.
(a) VEEV nsP2 protease/nsP23 substrate complex with P3 alanine (Ala1327). S3 residues Met702 and Ile698 are highlighted. (b) Complex as modeled in the Aura virus with a P3 valine, threonine at position 702, and glycine at position 698. (c) Complex as modeled in the salmonid alphaviruses (salmon pancreas disease virus and sleeping disease virus) with P3 valine, alanine at position 702, and valine at position 698. Substrate peptide is shown as sticks. Carbon atoms are green, nitrogen blue, and oxygen red. The nsP2 protease surface is shown with the N-terminal domain colored gray and the C-terminal domain colored tan. S3 subsite residues exhibiting significant variation are highlighted in magenta. All residue numbers correspond to VEEV numbering convention.