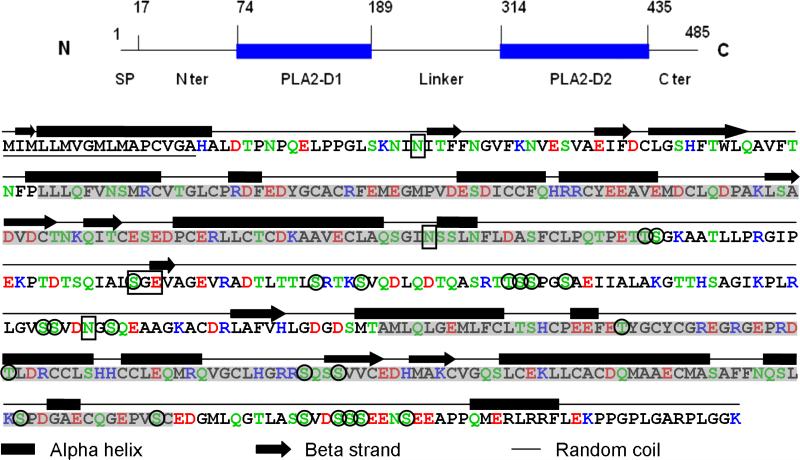
Figure 5. Primary and secondary structure of mouse OC90.
The schematic on the top is intended to assist the reader to navigate through the detailed molecule below. The N-terminal 17 amino acid signal peptide is underscored. The two sPLA2-like domains are shadowed. Acidic amino acids: red; basic amino acids: blue; hydrogen-bonding amino acids: green. A 25 amino acids hydrophilic stretch formed by TLTTLSRTKSVQDLQDTQASRTTSS is present in the linker segment. The secondary structure is predicted by the PSIPRED program (McGuffin et al., 2000). The three Asn sites for N-glycosylation predicted by the NetNGlyc 1.0 server, and the chondroitin sulfate site (SGE) predicted by Dr. Lijuan Zhang (pers. comm.) are boxed. The 23 phosphorylation sites suggested by NetPhos 2.0 prediction server are circled.