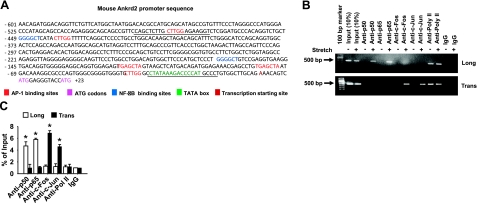
Figure 2.
Stretch activates Ankrd2 promoter through AP-1 and NF-κB transcription factors. A) Sequence of Ankrd2 promoter shows putative binding sites for AP-1 (red) and NF-κB (blue) transcription factors followed by a TATA box (green), transcription initiation site (bold black) and two ATG start codons (indigo). Underscores represent primer sequences used in ChIP assays. B) Diaphragm of WT mouse was excised and stretched either longitudinally or transversely to the myofibers for 15 min or maintained under unstretched conditions. Immediately after stretch, the diaphragms were cross-linked with formaldehyde and processed for ChIP assays using c-Fos, c-Jun, p50, p65, RNA polymerase II (positive control), or nonspecific antibody (negative control). Equal amount of resulting DNA (100 ng) from the ChIP assays were subjected to PCR analyses with primers specific for Ankrd2 or GAPDH promoter and resolved in 2% agarose gel to show the binding activity of AP-1 or NF-κB on the Ankrd2 promoter. The fraction of chromatin used in the ChIP is shown (10% input). C) Equal amounts of DNA from the above samples were subjected to real-time qPCR assays to estimate Ankrd2 or GAPDH promoter DNA levels. The ΔCT value of Ankrd2 DNA level was calibrated against that of the input DNA level to obtain the ΔΔCT value. A 1.0-fold change in site occupancy indicates no difference in the global transcription or IP efficiency. Values represent the mean ± se of 3 independent experiments. *P < 0.05 for corresponding stretch and nonstretch samples. Gel pictures are representative of 3 independent experiments (n=3).