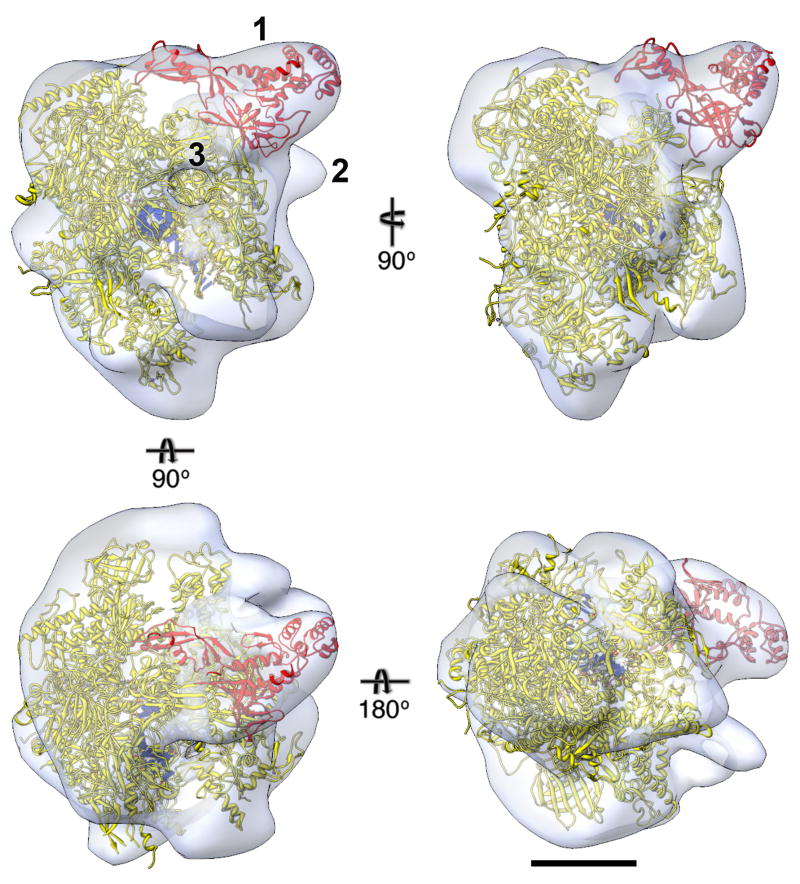
Figure 3. EM density map of human RNAP II with docked crystal structure.
Different views of the final density map (white translucent surface) of human RNAP II filtered to 25 Å resolution calculated from 22,704 particles, into which the crystal structure of yeast RNAP II was placed (Rpb4/7 in red, other subunits in yellow, nucleic acid in blue; PDB entry 1Y77 19). Label “1” indicates the region of the density map into which the atomic model of Rpb4/7 was moved. Label “2” indicates density not accounted for by the crystal structure, which may represent the C-terminal domain of Rpb1 and/or part of the antibody against Rpb1 used to recruit RNAP II to the Affinity Grid. Label “3” indicates the exit channel for the newly synthesized RNA. Scale bar is 5 nm.