Force-independent part of potential (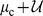 ) ) |
|
J |
Models no growth for vanishing force. |
Temperature (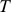 ) ) |
|
K |
— |
Actin stress fiber angle (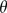 ) ) |
|
degrees |
Effect of varying 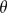 can be subsumed by varying can be subsumed by varying 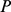 . . |
Young's modulus (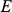 ) ) |
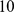
|
kPa |
Estimate for soft, gel-like biological materials. |
Bending modulus (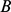 ) ) |
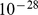
|
N.m
|
Estim. from 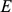 and cell memb. thickness and cell memb. thickness 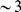 nm. nm. |
Cell memb. curvature ( ) ) |
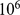
|
m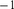
|
Estim. from cell height 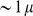 m. m. |
Initial FA length (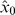 ) ) |
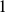
|
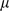 m m |
Typical focal complex length. |
FA width (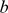 ) ) |
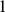
|
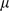 m m |
Estim. from images in [4] and [2]. |
Upper bound on complex size (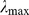 ) ) |
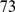
|
nm |
Motivated by [29]. |
Lower bound on complex size (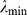 ) ) |
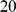
|
nm |
Integrin packing density; see [30]. |
Conformational displacement (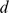 ) ) |

|
nm |
Estimated from integrin domain size. |
Kinetic coeff. (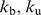 ) ) |
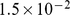
|
s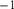
|
For sliding velocities 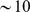 nm nm s s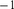 ; see [2]. ; see [2]. |
Ref. displacement for unbinding ( ) ) |
1.48 |
nm |
From ref. [28]. |
Max. conc. (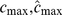 ) ) |
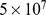
|
m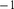
|
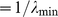 . . |
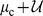 )
)
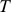 )
)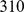
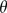 )
)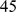
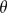 can be subsumed by varying
can be subsumed by varying 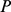 .
.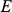 )
)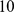
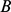 )
)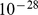

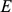 and cell memb. thickness
and cell memb. thickness 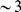 nm.
nm. )
)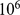
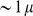 m.
m. )
)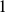
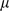 m
m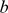 )
)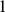
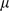 m
m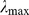 )
)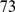
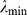 )
)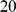
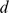 )
)
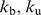 )
)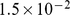
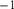
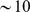 nm
nm s
s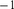 ; see [2].
; see [2]. )
)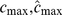 )
)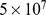
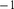
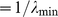 .
.