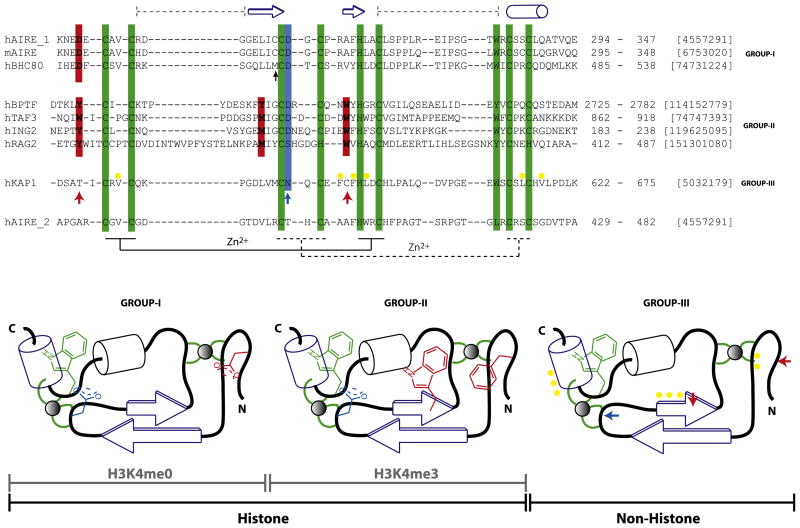
Figure 4. Classification of the PHD Finger Family.
Sequence features of structurally characterized three distinct subclasses of the PHD finger family with respect to ligand binding specificity, i.e., H3K4me0 (group I), H3K4me3 (group II), and nonhistone binding (group III). The recognition of H3K4me3 takes place by embracement of the trimethyl groups by characteristically positioned aromatic cage residues, whereas that of K4me0 is due to an ion pair formed with a distinct N-terminal Asp residue. The characteristic histone-peptide interacting positions are in red, and the Zn-chelating residues (the first and second tetrads are connected by regular and dotted lines, respectively) and conserved C-terminal aromatic residue characteristic of the entire PHD family are in green. In either of the H3 interacting PHD fingers, H3R2 often interacts with Asp/Glu (blue). These “red” and “blue” positions are absent in KAP1-PHD finger (bottom) indicated by ↑ that binds the adjacent bromodomain’s ZA helix by patch of nonpolar residues (yellow). Topology diagrams (bottom; based on Aravind et al. [2006]), not drawn to scale, highlight these features for clarity. The domain boundaries and gi numbers are indicated in the alignments. The secondary structural elements of the AIRE-PHD1 are indicated above the sequence. The black ↑ indicates position where similar interactions involving protein side chain is observed in AIRE and BHC80 PHD fingers. The sequence of AIRE-PHD2 is shown below the alignment to show its grouping with group III as non-H3 binder.