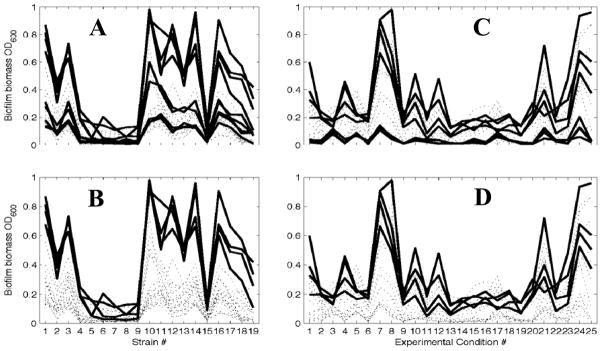
Fig. 2. Outcome of the computational analysis of the high-throughput experiment.
For A and B, quantitative amounts of biofilm were plotted versus the bacterial strains (labels on x-axis correspond to strain designations in column 1 of Table 1). Each line represents one experiment that is defined through a combination of four individual conditions. The peak at strain 10 is the ackA mutant at 37°C in TSB. Fig. 2A highlights (bold lines) the condition 37°C (p-value of 6.991 × 10−4), while Fig. 2B highlights the combination of the conditions 37°C as the growth temperature and TSB as the culture medium (p-value of 5.1558 × 10−47). Both panels show the entire data set, and only differ in their highlighting.
For Figs. 2C and 2D, the quantitative amounts of biofilm were plotted versus combinations of experimental conditions (labels on x-axis correspond to the designations in column 1 of Table 2). Each line represents one mutant. The peak at conditions 7 and 8 is at 37°C in TSB, the highest biofilm amount is formed by the ackA mutant (see A and B). Fig. 2C highlights mutants that lack genes with GO:0042867, pyruvate catabolic process function. Fig. 2D highlights mutants that lack genes that together show the presence of GO:0042867 and the absence of GO:0007155, which is characteristic of the type I fimbrium. Note that while the filters were defined requiring GO:0042867 to be present and GO:0007155 to be absent, other GO terms are associated with the same filters: For example, GO:0009289 and GO:0042995 select the same genes as GO:0007155 because they are all associated with type 1 fimbriae. Likewise, GO:0005624, GO:0006082, GO:0008776, GO:0009063, GO:0016052, GO:0016774, and GO:0045733 select the same genes as GO:0042867 because they are all associated with the same enzymes. For a complete of all GOs that were associate with a particular protein function, please, see Supplemental Table 1.