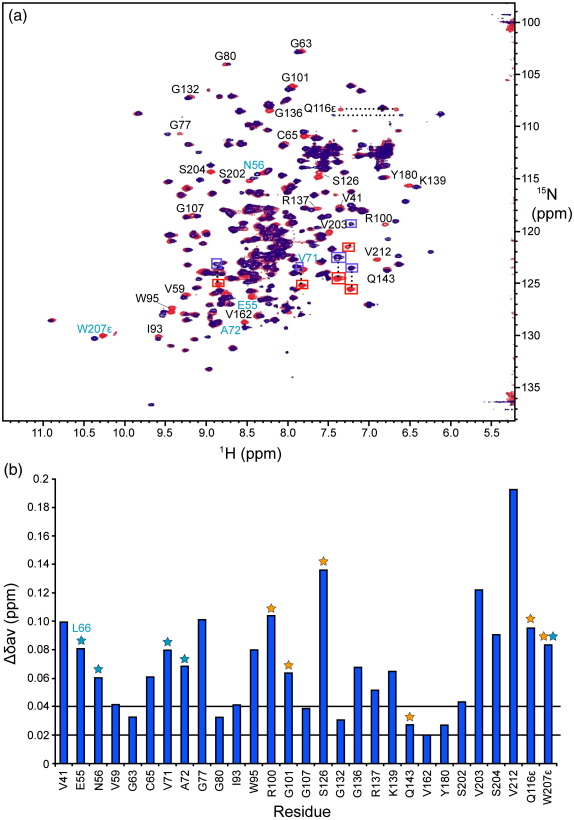
Fig. 2.
Identification of key interacting residues of mSPSB2. (a) Comparison of HSQC spectra of 0.1 mM uniformly N-labeled mSPSB2(12–224), free (red) and in a 1:1 complex with hPar-4(59–77) (blue). Spectra were recorded in 95% H2O/5% 2H2O, pH 6.7, 295 K, at 500 MHz. The red peaks are labeled with sequence-specific assignments for free mSPSB2(12–224) using the one-letter code and sequence positions (black). Conserved residues of GUSTAVUS and three SPSB proteins that are important for GUSTAVUS/VASA10 and mSPSB2/hPar-45 interactions are represented in cyan. Aliased resonances arising from Arg side chains of mSPSB2 are shown in red (free) and blue (complex) square boxes as these were due to different spectra widths used in the 15N dimension. (b) Weighted average chemical shift variations of 15N and 1H between free and bound forms of 15N-labeled mSPSB2(12–224). Cyan asterisks represent those residues that are conserved in GUSTAVUS, mSPSB1, mSPSB2, and mSPSB4 and have been shown to be involved in GUSTAVUS binding to VASA,10 except for mSPSB2 E55, where L66 is found in GUSTAVUS, mSPSB1, and mSPSB4. Orange asterisks indicate those residues that are known to be involved in mSPSB2/hPar-4 interaction according to a previous study.5 Horizontal lines show cutoffs for weighted average chemical shift differences of 0.02 and 0.04 ppm. The Trp207 peak is from the indole NH; the backbone amide resonance for this residue is close to the water resonance5,21 and difficult to follow.