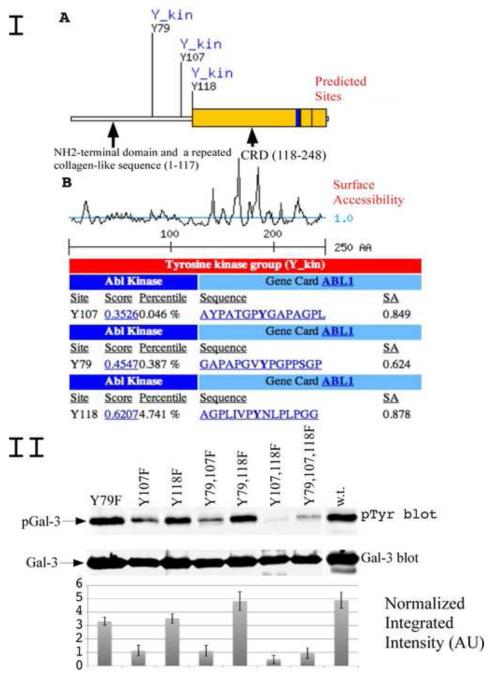
Figure1.
I. Bioinformatic search predicting possible tyrosine phosphorylation sities of galectin-3 and the cognate protein kinase. (A) A diagram depicting newly identified galectin-3 phosphorylation sites and potential kinase. Indicated are NH2-terminal domain, a repeated collagen-like sequence and carbohydrate binding domain (CRD).
(B) A diagram showing surface accessibility, scores and amino acids around predicted phosphorylation sites.
II. c-Abl phosphorylation of galectin-3 in vitro. Recombinant GST-Galectin-3 mutants of predicted tyrosine sites and galectin-3 wild type were used as substrate for active c-Abl in in vitro assay. The reaction was stopped by adding sample buffer, resolved on 10% polyacrilomide gel, and immunoblotted using anti-pTyr antibody (top) or anti-galectin-3 antibody (bottom). The normalized integrated intensity was calculated as Band integrated intensity/Line normalization factor, anti-galectin-3 blot was used as reference channel.