Abstract
We examined the expression of Nicotiana attenuata (Na) and Nicotiana obtusifolia (No) herbivore-induced genes in synthetic autopolyploids (NaT and NoT) and five independent allopolyploid Nicotiana × obtusiata (N×o) lines to understand how the expression of genes regulating complex polygenetic defense traits is altered in the early stages of allopolyploid hybridization. In Na, applying Manduca sexta oral secretions (OS) to wounds rapidly increased the transcript accumulation of wound-induced protein kinase (WIPK), lipoxygenase 3 (LOX3), nonexpressor of pathogenesis-related 1 (NPR1), and jasmonate-resistant 4 (JAR4) genes; these were correlated with increases in accumulation of jasmonic acid (JA), jasmonate-isoleucine, and trypsin protease inhibitors (TPIs). In No, OS elicitation reduced NPR1 transcripts and increased the level of salicylic acid (SA) that appeared to antagonize JA and JA-mediated defenses. OS elicited N×o lines, accumulated high levels of the uniparental transcript of WIPK, LOX3, JAR4, and TPI, but low levels of both parental NPR1 transcripts that in turn were correlated with an increase in SA and a decrease in JA levels, suggesting SA/JA antagonism in the allopolyploid crosses. Methyl jasmonate treatment of N×o lines elicited transcripts of both parental LOX3, JAR4, and TPIs, demonstrating that the uniparental pattern observed after OS elicitation was not due to gene inactivation. TPIs were induced at different levels among N×o lines; some lines expressed high levels comparable to Na, others low levels similar to No, suggesting that synthetic neoallopolyploids rapidly readjust the expression of their parental defensive genes to generate diverse antiherbivore responses. Changes in the expression of key genes and posttranscriptional events likely facilitate adaptive radiations during allopolyploid speciation events.
Polyploidy is an ongoing evolutionary process that generates new plant species. Estimates suggest that polyploidy is responsible for 2% to 4% of speciation events in angiosperms and 7% in ferns (Otto and Whitton, 2000; Blanc and Wolfe, 2004). Many of our cultivated plants are derived from autopolyploidy (duplication of single genome), for example, alfalfa (Medicago sativa) and potato (Solanum tuberosum), or from allopolyploidy (union of distinct genomes), for example, wheat (Triticum aestivum), oat (Avena sativa), cotton (Gossypium hirsutum), coffee (Coffea arabica), and canola (Brassica napus; Wendel, 2000). Other crops such as corn (Zea mays; Gaut and Doebley, 1997), soybean (Glycine max; Shoemaker et al., 1996), and cabbage (Brassica capitata; Lagercrantz and Lydiate, 1996) appear to have experienced polyploidization events in their evolutionary histories and are referred to as paleopolyploids. The model plant Arabidopsis (Arabidopsis thaliana), a typical diploid, is now considered as a paleopolyploid (Arabidopsis Genome Initiative, 2000). The widespread occurrence of polyploids presumably reflects their greater ability to evolve and adapt to new environments compared to their parental taxa. Allopolyploid lineages appear to undergo adaptive radiations more frequently than autopolyploid lineages. Indeed, by fusing two distinct genomes, allopolyploidy facilitates heterosis that offers a greater potential for adaptive diversification (Adams and Wendel, 2005; Flagel et al., 2008); nevertheless, the success of allopolyploids is unexpected given that the neospecies must be able to survive to the genomic shock associated with the fusion of two distinct genomes and to coherently express the parental polygenetic traits.
The last decade witnessed impressive advances in polyploidy research, with new results from Arabidopsis (Comai et al., 2000; Pontes et al., 2004), Brassica species (Lukens et al., 2006; Gaeta et al., 2007), cotton (Jiang et al., 1998), wheat (Han et al., 2003), and Nicotiana species (Lim et al., 2006; Pearse et al., 2006; Anssour et al., 2009). Most of these studies have focused on the genetic and genomic changes associated with the formation of polyploids, especially changes directly altering gene expression patterns, such as rapid and nonrandom changes including sequence elimination, changes in the DNA loci, and intergenomic cross talk, for example, transposon activation, chromosomal rearrangements, and chromosomal breaks (Kenton et al., 1993; Kitamura et al., 1997; Chase et al., 2003; Lim et al., 2004; Leitch et al., 2008). Recently, the dynamic changes affecting parental DNA sequences have received attention. Using Zingeria and Tragopogon species, both Kotseruba et al. (2003) and Lim et al. (2008) demonstrated that DNA sequence elimination may target only one or the other progenitor. In addition, using Gossypium allopolyploids, Adams et al. (2004) showed that the epigenetic silencing of parental homologous genes might be developmentally regulated, with one homolog silenced in some organs and the other silenced in other organs. These studies have provided mechanisms for the genetic changes that occur during polyploidy and specifically how the expression of one or the other parental genes may be modified; however, how these changes in parental gene expression influence the expression of functioning physiological systems that allow the hybrids to respond to their natural environment is still not clear. It is commonly assumed that the responses of most neoallopolyploids will be dysfunctional, and these dysfunctional hybrids are rapidly removed by natural selection. Hence, the adaptive radiation of polyploidy lineages is thought to be the result of the rapid winnowing of the bursts of genetic and functional diversity that results early in the neopolyploidization process. This assumption has not been thoroughly tested in any system.
The genus Nicotiana has many advantages for the study of polyploidization, not only because of its robust phylogenetic framework—it contains 75 species (Chase et al., 2003; Clarkson et al., 2004), 35 of which are recognized as allopolyploids (Clarkson et al., 2004; Leitch et al., 2008)—but also for its well-known ecology. Nicotiana quadrivalvis (Nq) and Nicotiana clevelandii (Nc), allopolyploids derived from amphidiploidy involving two diploid ancestors, Nicotiana attenuata (Na; as the paternal donor) and Nicotiana obtusifolia (No; as the maternal donor) approximately 2 million years ago (Chase et al., 2003), have been particularly useful for understanding how complex polygenic traits evolve. Lou and Baldwin (2003) reported that Nq and Nc retained different components of Na’s jasmonic acid (JA)-mediated defense response to attack from Manduca sexta larvae, most of which are fully mimicked by applying M. sexta oral secretions (OS) to wounds (Fig. 1; Wu and Baldwin, 2009). Most aspects of Na’s recognition response were retained with modifications in Nq, but many have been lost in Nc. Wu et al. (2006) demonstrated that maternally inherited (No) trypsin protease inhibitor (TPI) genes, which encode for protease inhibitors that reduce M. sexta performance, were retained in both Nc and Nq, whereas paternally inherited (Na) TPI genes were deleted. However, when these changes occurred, either directly after polyploidization or during the intervening 2 million years in either parents or allopolyploids, remains a mystery. Answering this question requires an understanding of the changes that occur rapidly after neopolypoloidization and can be addressed by comparing responses in both parental lines and newly resynthesized allopolyploids.
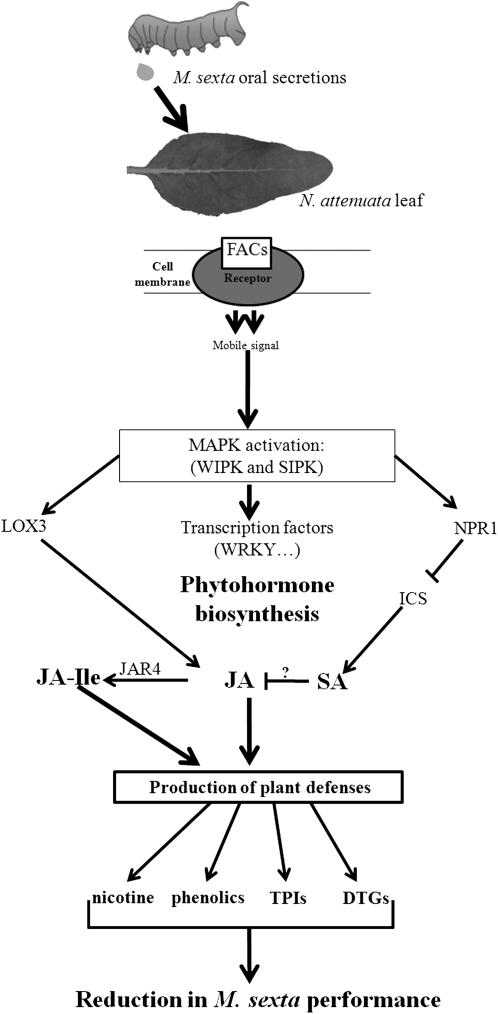
Figure 1.
An overview of the OS-elicited signaling cascade that activates direct defenses in Na’s leaves depicting the elements that were studied in the synthesized tetraploid and allopolyploid Nicotiana species. During herbivore attack by M. sexta larvae, FACs from larval OS bind to hypothetical receptors in the cell membrane at the attack site and activate unknown short-distance mobile signals. These signals activate mitogen-activated protein kinases that include SA-induced protein kinases and WIPKs that phosphorylate transcription factors (such as WRKYs), which in turn activate phytohormone signaling such as JA, SA, and JA-Ile and their associated biosynthetic genes such as LOX3, isochorismate synthase (ICS), and JAR4. By inhibiting ICS, NPR1 negatively regulates SA production and thereby SA/JA antagonism, allowing the expression of JA-mediated direct defenses such as nicotine, phenolics, TPIs, and diterpene glycosides (DTGs) that diminish the performance of M. sexta larvae feeding on elicited plants.
The first attempts, carried out by Pearse et al. (2006), to synthesize Na and No allopoylploids were unsuccessful, thus the authors used Nicotiana miersii as a maternal surrogate for No (the species most phylogenetically related to No) to create the synthetic neoallopolyploid, Nicotiana × mierata. By eliciting plants with OS and methyl jasmonate (MeJA) and comparing the changes in TPI activity, secondary metabolites, and released volatile organic compounds in the parents with those of the neoallopolyploid lines, the authors concluded that parental signaling cascades eliciting these defense responses had been reshuffled in the neopolyploids in a plug-and-play fashion to allow different secondary metabolite responses to be elicited by the diversity of OS- and JA-elicited signaling systems found in the parents. The analysis of metabolic responses provided by this study would benefit from information about how the expression of parental genes is altered in the synthetic polyploids, specifically, which parent genetically dominates and how this influences the defensive response of the synthetic allopolyploids.
Here we examine the changes in Na and No antiherbivore gene expression, phytohormone accumulation, and TPI activity in five independent lines of the allotetraploid Nicotiana × obtusiata (N×o) (Na [as the paternal donor] × No [as the maternal donor]), and autotetraploids of Na (NaT) and No (NoT) characterized by Anssour et al. (2009). We first dissect the specific transcript accumulation (levels and timing) of some of the early antiherbivore responsive genes in the OS-elicited signaling cascade (Fig. 1), namely, wound-induced protein kinase (WIPK), lipoxygenase 3 (LOX3), nonexpressor of pathogenesis-related 1 (NPR1), and jasmonate-resistant 4 (JAR4) of Na and No. Then, we measure the phytohormone levels of JA, jasmonate-Ile (JA-Ile), and salicylic acid (SA); finally, we analyze the kinetics of parental TPI transcript accumulation and TPI activity in the synthetic polyploids. Variations in the expression of antiherbivore signaling and resistance genes and in the levels of phytohormones and TPI activity among the synthetic polyploid lines are compared to their parental lines.
RESULTS
OS Elicitation Induces High Levels of Na-WIPK and No-LOX3 Transcripts and Attenuated Levels of Na- and No-NPR1 Transcripts in N×o Lines
Na’s antiherbivore response is rapidly initiated after the introduction of M. sexta OS into wounds. This defensive response is highly specific and its activation depends on the plant's ability to recognize fatty acid-amino acid conjugates (FACs) present in M. sexta OS. Recently, Wu et al. (2007) demonstrated that FACs rapidly activate two mitogen-activated protein kinases, WIPK and SA-induced protein kinase, which in turn stimulate the expression of NPR1 and JA biosynthetic genes, such as LOX3 (Fig. 1). To understand how the expression of these early antiherbivore responsive genes are altered after synthetic auto- and allopolyploidy, we measured the transcript accumulation of WIPK, LOX3, and NPR1 in the diploid and the synthetic polyploid lines subjected to wounding and OS elicitation performed on the +1 leaves, leaves which had just completed the source-sink transition.
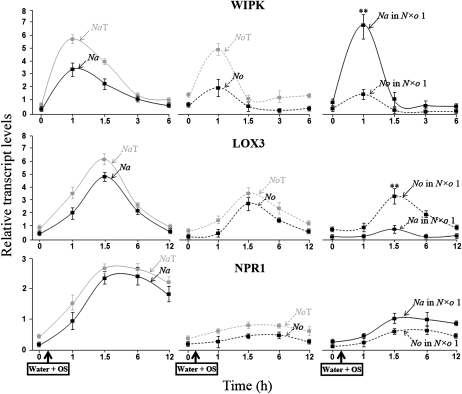
Na and No responded differently to OS elicitation; Na dramatically increased the transcript levels of WIPK (reaching a maximum at 1 h after OS elicitation), LOX3 (a maximum level at 1.5 h), and NPR1 (a maximum level at 2.5 h), whereas No accumulated much lower levels of these transcripts with similar patterns as in Na. The autotetraploids, NaT and NoT, followed a similar pattern of WIPK, LOX3, and NPR1 transcript accumulations and did not differ significantly compared to their respective diploids. After OS elicitation, the allopolyploid N×o lines induced both parental WIPK and NPR1 transcripts, but showed uniparental accumulation for LOX3 transcripts; in this concern all N×o lines expressed No-LOX3 transcripts, but did not induce any Na-LOX3 transcripts.
To get an insight into the genetic interactions acting on the expression of LOX3, WIPK, and NPR1 genes, we conducted a graphical analysis described by Zhang and Borevitz (2009). In this model, the authors suggest that the expression of a given gene is under the control of only cis-genetic elements, if the specific parental expression difference is equal to the allele expression difference in the hybrid. Otherwise, both cis- and trans-elements are involved as the parental expression difference can be explained by cis-effect (the allele expression difference in the hybrid), plus composite trans-effect. A similar analysis comparing parental transcript accumulations among the auto- and allopolyploids, elicited by M. sexta OS, revealed that both cis- and trans-regulatory elements act on the expression of WIPK, LOX3, and NPR1 genes (Supplemental Fig. S2).
In short, Na and No responded differently to OS elicitation; Na dramatically increased the transcript levels of WIPK, LOX3, and NPR1, whereas No accumulated much lower levels.
In the autopolyploids, the transcript accumulation of WIPK, LOX3, and NPR1 followed a pattern similar to that observed in the parental lines. All allopolyploid lines favored the accumulation of Na-WIPK and No-LOX3 transcripts. Finally, we suggest that both cis- and trans-regulatory elements act on the expression of WIPK, LOX3, and NPR1 gene in the allopolyploid lines.
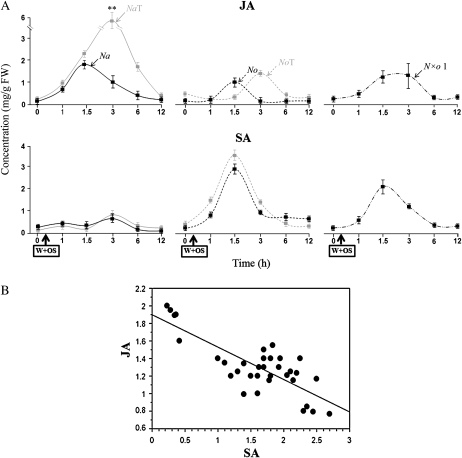
The Patterns of OS-Elicited SA and JA Accumulation Suggest JA/SA Antagonism in the Allopolyploid Lines
OS elicitation results in phytohormone bursts that spread throughout the attacked leaf to trigger defense responses, which are tailored by interactions among different phytohormones. In Na, M. sexta OS elicits a JA burst, and a much more modest response in SA levels, which is known to be down-regulated by an OS-elicited ethylene burst (Diezel et al., 2009) and the activity of NPR1 (Rayapuram and Baldwin, 2007). Hence, without the ethylene burst and the activity of NPR1, OS elicitation in Na would result in a large SA burst, which antagonizes the JA burst and attenuates the JA-elicited defense responses (Diezel et al., 2009). In contrast to Na, No responds to OS elicitation with a large SA burst and an attenuated JA burst. To understand how synthetic polyploidy alters the accumulation of phytohormones during herbivory, we measured the levels of JA and SA after OS elicitation in the synthetic polyploids, and compared them to that of the parental diploids.
In Na and NaT, JA dramatically increased after OS elicitation, attaining maximum levels at 1.5 and 3 h, respectively. The maximum level of JA in NaT was three times that in Na. In No, NoT, and N×o lines, OS elicitation induced only low levels of JA; the highest level was about half as much as that in Na. Compared to the diploid lines, both auto- and allopolyploids were delayed in attaining maximum JA levels, as was observed in previous studies (Lou and Baldwin, 2003; Pearse et al., 2006; Wu et al., 2006).
In Na and NaT, OS elicitation induced attenuated levels of SA, whereas in No, NoT, and N×o lines, it induced high levels of SA, which reached a maximum 1 h after elicitation. The highest level of SA in No, NoT, and N×o lines was about 4 times that in Na. Statistical comparisons of JA and SA levels in Na, No, and N×o lines showed that JA is negatively correlated with SA (r = −0.683, P < 0.001; Fig. 3; Supplemental Fig. S3).
Figure 3.
Patterns of OS-elicited SA and JA accumulation reveal SA/JA antagonism in the allopolyploid lines. A, OS elicitation in Na and Na tetraploid (NaT: left sections) elicited strong JA bursts and attenuated SA bursts. In contrast, OS elicitation of No, No tetraploid (NoT; center sections), and N×o line 1 (N×o 1; right sections) produced small JA bursts, but large SA bursts. The JA burst was delayed in all tetraploid lines in comparison to those of the diploids. B, Correlations among levels of JA and SA in Na, No, and N×o lines revealed that JA levels are negatively correlated with SA levels (r = −0.68, P < 0.01). The measurements of JA and SA levels were performed on leaves elicited with 20 μL of deionized water mixed with 20 μL of M. sexta OS, and harvested at the indicated times. Asterisks indicate levels of significant difference between the maximum values of the pairs plotted together on the same graph (**P < 0.01).
In short, OS elicitation resulted in a JA burst in Na but an SA burst in No, and the allopolyploids followed a pattern similar to that observed in No. Only Na autopolyploids show a dosage-dependent increase in OS-elicited JA. Correlations among levels of JA and SA in Na, No, and N×o lines suggest SA/JA antagonism in the allopolyploid crosses.
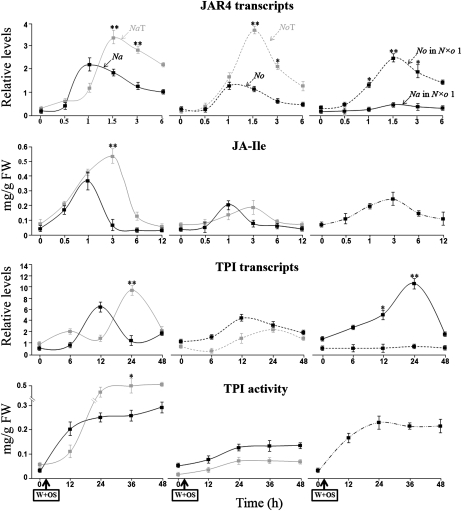
N×o Lines Enhance the Accumulation of Only One Parental Transcript of JAR4 and TPIs, and Accumulate Variable Levels of Active TPIs after OS Elicitation
We examined the transcript accumulation of JAR4 and TPIs as well as TPI activity levels in Na, No, and synthetic polyploids, after plants were induced with OS, to investigate changes caused by synthetic polyploidy in the transcript accumulation of OS-induced resistance genes, and the consequences of these changes for the expression of an important resistance trait.
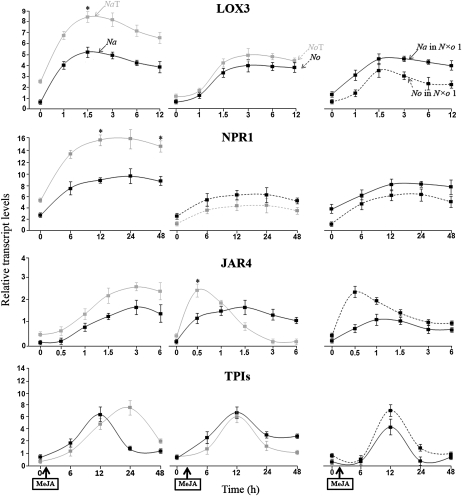
In Na and No, JAR4 transcript levels rapidly accumulated in response to OS elicitation; after 1 h, this level had reached a maximum and was higher in Na than in No. NaT and NoT showed a dosage-dependent increase in the levels of JAR4 compared to the levels in their respective diploids. N×o lines accumulated No-JAR4, but not Na-JAR4. Both auto- and allopolyploids showed a delay of 0.5 h in attaining maximum levels of JAR4 transcripts compared to the diploid lines.
JAR4 activity mediates the conjugation of Ile with JA to form JA-Ile, which in turn interacts with the F-box protein, COI, to mediate JA-dependent defenses. In Na and NaT, JA-Ile increased after OS elicitation, attaining maximum at 1.5 and 3 h, respectively. The maximum level of JA-Ile in NaT was 1.5 times that in Na. In No, NoT, and N×o lines, OS elicitation induced only attenuated levels of JA-Ile; the highest level was about 30% of the levels found in Na.
In Na and No, TPI transcripts increased after OS elicitation reaching a maximum after 12 h. TPI transcripts showed a dosage-dependent increase in NaT compared to Na, whereas in NoT the accumulation of TPI transcripts was reduced. N×o lines accumulated Na-TPIs, but not No-TPIs. Both No and N×o lines showed a delay of 12 h in reaching the maximum level of accumulated TPI transcripts compared to the diploid lines.
In Na and No, TPI activity increased after OS elicitation, reaching a maximum after 24 h; this level was higher in Na than in No. In NaT, but not in NoT, TPI activity showed a 2-fold dosage-dependent increase compared to that in the diploids. TPI activity in lines 1 and 2 were comparable to that of Na, while in lines 3 to 5, TPI activity was at basal levels comparable to that in No (Fig. 4; Supplemental Fig. S4).
Figure 4.
OS elicitation enhances the accumulation of No-JAR4 and Na-TPI transcripts, and increases the levels of JA-Ile and TPI activity in the allopolyploid lines. After elicitation with M. sexta OS, Na and No rapidly accumulate JAR4 transcripts and JA-Ile levels, as well as transcripts and levels of active TPIs. However, the levels of JAR4 and TPI transcripts, JA-Ile, and TPI activity were higher in Na than in No. Autopolyploidy resulted in increased accumulations of OS-elicited JAR4 transcripts, JA-Ile levels, transcripts, and TPI activity levels in Na, but not in No with the exception of No-JAR4 transcripts (which were more than double in the autopolyploid compared to the diploid No). N×o line 1 (N×o 1) accumulated only No-JAR4 and Na-TPI transcripts, but not Na-JAR4 and No-TPI transcripts. All polyploid lines showed a delay in the accumulation of JAR4, JA-Ile, as well as transcripts and TPI activity. Asterisks indicate levels of significant difference between the maximum values of the pairs plotted together on the same graph (*P < 0.05; **P < 0.01).
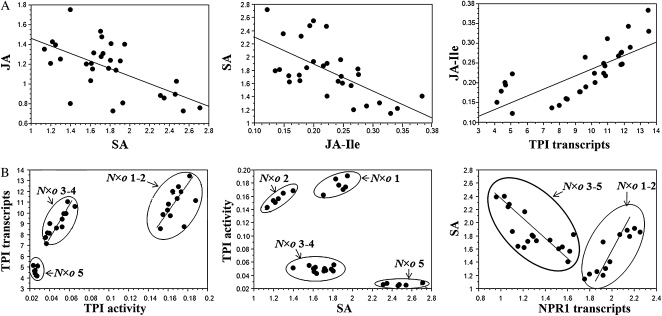
Statistical comparisons among levels of JA, SA, JA-Ile, and TPI transcripts accumulated in the allopolyploid lines, revealed that as in Na, SA was negatively correlated with JA (r = −0.608, P < 0.001), and JA-Ile (r = −0.631, P = 0.002), and JA-Ile was positively correlated with TPI transcript levels (r = 0.746, P < 0.001; Fig. 5). However, comparisons among transcripts and TPI activity levels showed different linear distributions among the allopolyploid lines that clustered in three distinct groups: N×o lines 1 and 2, N×o lines 3 and 4, and N×o line 5. Comparisons among levels of TPI activity and SA revealed that differences in SA influence TPI activity; N×o lines 1 and 2 were distributed in distinct groups based on differences in SA levels. Finally, comparisons among levels of SA and NPR1 transcripts suggest that N×o 1 and 2 and N×o 3 and 5 may react differently to variation in NPR1 transcript levels; while in N×o lines 3 to 5, SA accumulation is down-regulated by NPR1, N×o lines 1 and 2 seem to have adapted a different mechanism, probably involving ethylene.
Figure 5.
Variations in phytohormone cross talk (SA/JA) influence the accumulation of TPI activity in the allopolyploid lines. A, Statistical comparisons among levels of JA, SA, JA-Ile, and TPI transcripts accumulated in N×o lines revealed that SA was negatively correlated with JA (r = −0.608, P < 0.001) and JA-Ile (r = −0.631, P = 0.002), and JA-Ile was positively correlated with TPI transcript levels (r = 0.746, P < 0.001). B, Statistical comparisons among levels of SA, transcripts, and active TPIs revealed that the variation in TPI activity among N×o lines are caused by differences in accumulated SA (which negatively regulates JA-Ile). The levels of SA are differently regulated in N×o lines 1 and 2 and 3 to 5; while in N×o 3 to 5, SA accumulation is down-regulated by NPR1, N×o lines 1 and 2 seem to be using a different mechanism, which we speculate may involve ethylene production.
In short, after OS elicitation, autopolyploidy induced a dosage-dependent increase in the accumulation of JAR4 transcripts in both Na and No; in Na, this dosage effect was correlated with an increase in JA-Ile levels, TPI transcripts, and TPI activity. Whereas in No, there was no gene dosage effect beyond the accumulation of JAR4 transcripts. The accumulation patterns of JA-Ile, TPI transcripts, and active TPIs were similar between diploid and autotetraploid lines. All allopolyploid lines accumulated No-JAR4 and Na-TPI transcripts and increased levels of JA-Ile and TPI activity in response to OS elicitation. In the allopolyploid lines, SA was negatively correlated with JA and JA-Ile, and JA-Ile was positively correlated with TPI transcript levels. The allopolyploids accumulated variable levels of active TPIs due to differences in SA levels.
MeJA Elicitation of N×o Lines Reveals That OS-Elicited Uniparental Gene Expression Is Not Due to Gene Inactivation
The accumulation of one or the other parental transcript—namely, Na-LOX3, No-JAR4, and Na-TPIs—in N×o lines after OS elicitation was particularly noteworthy. To test the hypothesis that this pattern of transcript accumulation was due to inactivation of the nonexpressed parental gene, we elicited plants with MeJA and measured transcript accumulation of both parental copies of the LOX3, JAR4, and TPI genes. In Na and No, MeJA treatment elicited changes in the timing and levels of accumulation of LOX3, NPR1, JAR4, and TPI transcripts that were comparable to those elicited by OS elicitation. In Na, autopolyploidy induced a gene dosage-dependent increase in the transcript accumulation levels of LOX3, NPR1, and JAR4, but not in the levels of TPIs. In NoT, JAR4 transcripts showed a gene dosage-dependent increase, but not those of TPIs and LOX3. Unlike OS elicitation, MeJA treatment of N×o lines induced the accumulation of No-LOX3, Na-JAR4, and No-TPI transcripts (Fig. 6; Supplemental Fig. S5), demonstrating that these parental gene copies are functional.
Figure 6.
MeJA treatment of N×o line 1 (N×o 1) elicits transcripts of both parental LOX3, JAR4, and TPIs, demonstrating that the uniparental pattern of transcript accumulation observed after OS elicitation is not due to gene inactivation. After MeJA application, N×o line 1 accumulated both parental LOX3, JAR4, and TPI transcripts with a similar pattern to that of the parental lines (Supplemental Fig. S3). Application of MeJA restored the delay in the accumulation of JAR4 and TPI transcripts observed after M. sexta OS elicitation. Asterisks indicate levels of significant difference between the maximum values of the pairs plotted together on the same graph (*P < 0.05; **P < 0.01).
In short, MeJA treatment resulted in a gene dosage-dependent increase in the accumulation of LOX3, NPR1, and JAR4 in NaT and in the accumulation of only JAR4 transcripts in NoT. Treatment of allopolyploid lines with MeJA elicited both Na and No LOX3, JAR4, and TPI transcripts, demonstrating that the uniparental pattern of these genes observed after OS elicitation was not due to gene inactivation.
DISCUSSION
Polyploidization is thought to provide evolutionary and ecological advantages to plant species over their parental taxa (Adams and Wendel, 2005). A commonly proposed explanation is that polyploidy, by increasing gene redundancy, promotes novel functions that allow neospecies to adapt to wide range of habitats, and survive under unfavorable conditions (Otto and Whitton, 2000; Soltis and Soltis, 2000). Studies suggest that the occurrence of new traits in allopolyploids might be the consequence of flexible integration of parental components. Recently, Lou and Baldwin (2003) and Pearse et al. (2006) demonstrated that parental defensive components are reshuffled among allopolyploid lines to generate diverse defensive responses against M. sexta attack. However, both of these studies lacked information on the genetic causes of the diversity observed in the defensive responses of the allopolyploids. Here, we investigate how parental antiherbivore gene expression is altered in synthetic autopolyploid of Na and No (NaT and NoT) and their allopolyploid lines N×o (lines 1–5) characterized by Anssour et al. (2009), and the consequences of the resulting alterations for plants’ defensive responses. The results demonstrate that allopolyploids rapidly generate variability in their antiherbivore defensive responses by altering the expression of particular parental components involved the herbivore recognition, phytohormone signaling, and resistance responses.
In Na, the antiherbivore defensive response is rapidly initiated after the introduction of FACs present in M. sexta OS to wounds. Wu et al. (2007) demonstrated that this initial recognition response is associated with a rapid accumulation of WIPK, LOX3, and pathogenesis-related (NPR1) gene transcripts. The comparison of WIPK, LOX3, and NPR1 transcript levels in Na and No revealed that these two species respond differently to OS elicitation; while Na dramatically increases the level of WIPK, LOX3, and NPR1, No accumulates reduced levels of WIPK, LOX3, and does not induce the accumulation of NPR1 transcripts. Allopolyploidy induces variability in the accumulation of all parental transcripts in N×o lines; some transcripts show an increase in their accumulation (Na-WIPK and No-LOX3), others a reduction (No-WIPK, Na-LOX3, Na and No NPR1; Fig. 2; Supplemental Fig. S1). These variations in the transcript accumulation of WIPK, LOX3, and NPR1 genes in the allopolyploids are probably a consequence of alterations in the regulatory network that controls the specific expression of these genes. Indeed, under OS elicitation, WIPK, LOX3, and NPR1 gene expression is under the control of cis- and trans-regulatory elements (Supplemental Fig. S2), both of which are known to be preferentially associated with epigenetic elements that repress and activate gene expression (Zhang and Borevitz, 2009). These regulatory elements are also known to alter gene expression in other allopolyploid and interspecific hybrid systems (Wang et al., 2004, 2006; Wittkopp et al., 2004; de Meaux et al., 2006; Stupar and Springer, 2006; Chen, 2007).
Figure 2.
OS elicitation induces uniparental transcript accumulation of WIPK and LOX3, but not NPR1, in the synthetic allopolyploids. After elicitation with M. sexta OS, Na dramatically increases the levels of WIPK, LOX3, and NPR1 transcripts, whereas No accumulates comparatively lower levels of WIPK, LOX3, and NPR1 transcripts. Autopolyploid Na and No show dosage-dependent increases in the accumulation of WIPK transcripts, but not in those of LOX3 and NPR1. The synthetic allopolyploid, N×o line 1 (N×o 1), exhibits uniparental patterns of transcript accumulation (of Na-WIPK and No-LOX3), and attenuated levels of Na-LOX3 and Na- and No-NPR1 transcripts. The transcript accumulation was analyzed by SYBR Green PCR. For this, single leaves from seven to eight replicate plants were wounded with a fabric pattern wheel and the wounds were immediately treated with 20 μL of deionized water mixed with 20 μL of M. sexta OS. Treated leaves were then harvested at the indicated times. All values were normalized to actin as an internal standard. Asterisks indicate levels of significant difference between the maximum values of the pairs plotted together on the same graph (*P < 0.05; **P < 0.01).
Herbivore attack results in phytohormone bursts that spread throughout the attacked leaf to trigger defense responses. In Na, M. sexta OS elicits a JA burst, and a much more modest response in SA levels, which is known to be down-regulated by an OS-elicited ethylene burst (Diezel et al., 2009) and the activity of NPR1 (Rayapuram and Baldwin, 2007). However, in No and the synthetic allopolyploids, OS elicitation induced attenuated levels of JA and a dramatic increase in SA levels, which seems to antagonize JA (Fig. 3; Supplemental Fig. S3). The elicited JA can be conjugated with various amino acids (Sembdner and Parthier, 1993; Sembdner et al., 1994). Recently, using Na, Kang and Baldwin (2006) demonstrated that JA conjugation with amino acids is mediated by JAR4 (the Arabidopsis JAR1 homolog), which adenylates JA before its conjugation. JA-Ile, the most abundant of the JA conjugates (Staswick et al., 2002; Staswick and Tiryaki, 2004), is considered as the principle phytohormone elicitor of TPI production in Na (Wang et al., 2007), and its accumulation facilitates the physical interaction between jasmonate ZIM domain and coronatine-insensitive proteins to increase downstream antiherbivore resistance response. In Na and No, M. sexta OS elicits a rapid accumulation of JAR4 transcripts, JA-Ile, as well as TPI (transcripts and activity) levels. This resistance response is more pronounced in Na than in No that accumulates low levels of JA. In the allopolyploid lines, OS elicitation induced the expression of only one parental transcript of JAR4 and TPIs. TPI activity (timing and levels) was variable among the allopolyploid lines; TPI activity in N×o lines 1 and 2 was comparable to that in Na, whereas TPI activity in lines 3 to 5 was comparable to that in No (Fig. 4; Supplemental Fig. S4).
It is not clear how the variability in the accumulated active TPIs is generated, but statistical comparisons among levels of JA, SA, JA-Ile transcripts, and TPI activity in the allopolyploid lines suggest that this variability is generated during the posttranslational modification of TPI expression. Indeed, comparisons among transcripts and TPI activity levels showed different linear distributions among allopolyploid lines that were separated in three distinct groups: N×o lines 1 and 2, N×o lines 3 and 4, and N×o line 5. Correlation analysis conducted on TPI activities and SA levels suggest that variations in TPI activity of N×o lines 1 and 2 and 3 to 5 are caused by differences in accumulated SA (Fig. 5). These results agree with previous finding, suggesting that SA might be involved in the processing and the maturation of protease inhibitors, by eliciting vacuolar proteases (Horn et al., 2005). The levels of SA appear to be differently regulated in N×o lines 1 and 2 and 3 to 5; while in N×o 3 to 5, SA accumulation is down-regulated by NPR1, N×o lines 1 and 2 seem to have adapted a different mechanism (Fig. 5), probably involving ethylene, known to down-regulate SA accumulation. Hence, variations in phytohormone cross talk (SA/JA and likely ethylene/SA) can account for much of the expressed TPI activity in the allopolyploid lines.
The accumulation of one or the other parental transcript—namely, Na-LOX3, No-JAR4, and Na-TPIs in N×o lines—in response to OS elicitation was of particular interest, since similar uniparental expression pattern of homologous genes have been reported in several studies using Arabidopsis (Chen, 2007), or Gossypium allopolyploids (Adams et al., 2004). Here, to understand this transcript accumulation pattern, we initially assumed that the nonexpressed parental gene copies in N×o lines had lost their functionality due either to chromosomal translocations (or transposition) or to DNA sequence elimination; all these genetic changes have frequently been reported in several allopolyploids (Song et al., 1995; Feldman et al., 1997; Shaked et al., 2001; Skalicka et al., 2005; Tate et al., 2006). However, in response to MeJA treatment, all N×o lines accumulated both parental LOX3, JAR4, and TPI transcripts (Fig. 6; Supplemental Fig. S5), suggesting that the uniparental transcript accumulation pattern observed after OS elicitation was not due to inactivation of LOX3, JAR4, and TPIs gene expression. An alternative hypothesis would be that some OS-elicited regulatory elements, influencing the expression of these genes, were lost through homologous recombinations during polyploidy.
Interestingly, unlike OS elicitation, MeJA treatment of the autopolyploids resulted in a gene dosage-dependent increase in the accumulation of LOX3 (in NaT and NoT), NPR1 (only in NaT), and JAR4 transcripts (only in NoT). This increase in transcript accumulation in the autopolyploids might also reflect an increase in expressed gene copies; suggesting that under OS elicitation, the expression of gene homologs in polyploids might be subjected to a selective mechanism that restricts gene expression to specific parental copies. Whether this regulatory mechanism is achieved by specific elements that differentially target one or the other parental copy, or via epigenetic modifications (e.g miRNA, histone methylation, or acetylation) is still not clear. Particularly, in N×o lines, the recovery of No-LOX3 transcript accumulations after MeJA elicitation is consistent with a regulatory block that discriminates between the parental LOX3 transcripts elicited by OS. Such a regulatory mechanism is probably located upstream of JA signaling.
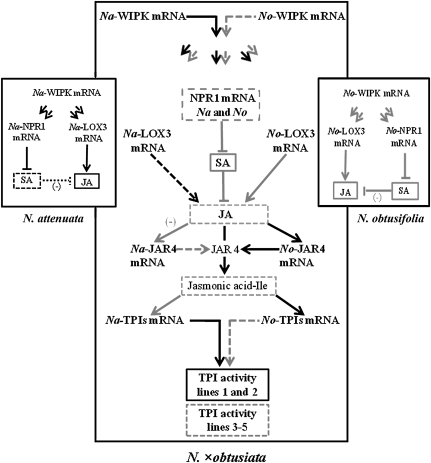
Synthetic auto- and allopolyploidy induced a reshuffling in the accumulation of parental defensive gene transcripts, phytohormones, and active TPI levels that mediate the antiherbivore resistance responses. Allopolyploidy seems to have integrated both the defensive components of Na and No. However, in response to OS elicitation, N×o lines accumulated only one or the other parental defensive transcripts and generated variability in expressed active TPI levels (Fig. 7). Here, we demonstrated that the variation in the level of active TPIs among the allopolyploid lines is probably a consequence of differences in phytohormone cross talk (SA/JA and likely ethylene/SA). We propose that the alterations in parental gene transcripts accumulated in N×o lines is the consequence of epigenetic and regulatory network changes that may facilitate adaptive radiations during allopolyploidy. More detailed molecular studies are required to understand the timing and frequency of these changes among natural polyploids.
Figure 7.
Schematic summary of the OS-elicited components of the signal cascade that elicits antiherbivore defense responses in the synthetic allopolyploid lines. After elicitation with M. sexta OS, N×o lines accumulate high levels of Na-WIPK, No-LOX3, No-JAR4, and Na-TPIs, but low levels of Na- and No-NPR1 transcripts. The low accumulated transcript levels of NPR1 were correlated with an increase in SA and a decrease in JA levels, suggesting SA/JA antagonism in N×o lines. TPI levels were variable among N×o lines; the patterns of TPIs accumulation in lines 1 and 2 were similar to that in Na, whereas that of lines (3–5) were comparable to that in No. This model suggests a rapid readjustment of the expression of Na and No defensive genes to generate a diversity of antiherbivore responses. Transcripts or metabolites that accumulated at low levels after OS elicitation are represented by dashed arrows and lines, while those that accumulated at high levels are represented by solid arrow and lines.
MATERIALS AND METHODS
Plant Material
Plant Breeding
Na’s seeds originated from a native population in Utah (Baldwin et al., 1994) and were inbred for 17 generations. No’s seeds were collected in 2004 at the Lytle ranch preserve (Santa Clara, UT) and inbred for one generation. The polyploids’ seeds, inbred for five generations, were produced from synthetic polyploids described by Anssour et al. (2009).
Plant Growth
Seeds from all studied species and lines were germinated and grown as described by Krügel et al. (2002). Briefly, seeds were treated with smoke before being sterilized for 1 h with 0.1 mm gibberellic acid and germinated on sterile agar with Gamborg B5 media (Duchefa). After 10 d of growth, seedlings were transferred to soil-based growth medium in Teku pots and, after an additional 10 d, transplanted to soil in 1-L individual pots and grown in a glasshouse at 26°C to 28°C under 16 h of light supplied by Philips Son-T Agro 400-W sodium lights. Plants in the rosette stage of growth were used in all experiments.
Plant Treatment and Sample Harvest
Plant treatments were conducted as described by Lou and Baldwin (2003) with some modifications. For W + OS treatments, +1 leaves from each species and lineage were damaged by rolling a fabric pattern wheel to create a standardized mechanical wound, then 20 μL of OS (1:1 diluted with deionized water) from fourth- to fifth-instar Manduca sexta larvae was rubbed into the wounds. For MeJA treatment, MeJA was dissolved in heat-liquefied lanolin at a concentration of 7.5 mg/mL; 20 μL of the resulting lanolin paste was applied to +1 leaves to elicit the plants with 150 μg of MeJA. Untreated control plants were used in every experiment. After specific times, leaves were excised, immediately frozen in liquid nitrogen, and stored at −80°C until analysis.
Protein Extraction and TPI Activity Assay
Leaf tissues from each species and lineage were induced with M. sexta OS or with MeJA (one of six or seven plants/species or lineage/time point), and prepared for a TPI quantification. Briefly, plant tissues (approximately 150 mg) were crushed in liquid nitrogen, and 500 μL extraction buffer (0.1 m Tris-HCl [pH = 7.6], 2 mm polyvinylpyrrolidone, 13 mm phenylthiourea, 30 mm diethyldithiocarbamate, 60 mm ethylene diamine tetraacetic acid) was added for every 100 mg of tissue. Leaf tissue was then completely suspended by vortexing. After being centrifuged at 4°C for 20 min, supernatant was transferred to a fresh tube. Total protein content in each sample was determined using a Bio-Rad protein assay kit (Bradford assay) against serial dilutions of bovine serum albumin as a standard. TPI activity was determined by radial diffusion activity as described by Van Dam et al. (2001).
Phytohormone Analysis Using Liquid Chromatography-Tandem Mass Spectrometry
For the phytohormone analysis, approximately 300 mg of crushed frozen leaf tissue sample from each species and lineage was transferred to a FastPrep tube containing 0.9 g of FastPrep matrix, 200 ng/mL of isotope labeled JA (1,2-13C-JA), and pCA (as an internal standard), as well as 1 mL of extraction buffer (acetone: 50 mm citric acid [7:3 v/v]). Samples were then homogenized for 45 s at a speed 6.5 in a FastPrep homogenizer (Thermo Electron, http://www.thermo.com) and afterward centrifuged at maximum speed (16,000g) for 10 min at 4°C. Supernatants were transferred to fresh tubes to be subsequently evaporated in a vacuum concentrator (Eppendorf, http://www.eppendorf.com) to remove the remaining traces of acetone, and then extracted twice with 2 mL of ether. The ether phases of each sample were evaporated to dryness in the vacuum concentrator; the pellets were suspended with 70% methanol and the phytohormone measurements were conducted on a liquid chromatography-tandem mass spectrometry system (Varian 1200; Varian, http://www.varianinc.com). Fifteen microliters of each sample was injected onto a ProntoSIL column (C18; 5 mm, 50 × 2 mm; Bischoff, www.bischoff-chrom.com) attached to a precolumn (C18, 4 × 2 mm; Phenomenex, www.phenomenex.com). The mobile phase consists of 0.05% formic acid (solvent A) and 0.05% formic acid in acetonitrile (solvent B) used in a gradient mode with the following conditions: time/concentration (min/%) for B: 0:00/15; 1:30/15; 4:30/98; 12:30/98; 13:30/15; 15:00/15 with a flow of (time/flow [min/mL]): 0:00/0.4; 1:00/0.4; 1:30/0.2; 10:00/0.2; 10:30/0.4; 12:30/0.4; 15:00/0.4. Compounds were detected in the electrospray ionization negative mode. Molecular ions [M-H](2) at mass-to-charge ratio (m/z) 137 and 209 and 141 and 213 generated from endogenous phytohomones and their internal standards, respectively, were fragmented under 15-V collision energy. The ratios of ion intensities of their respective daughter ions, m/z 93 and 97 and m/z 59 and 63, were used to quantify endogenous phytohomones.
Molecular Cloning
cDNA molecular cloning and sequencing was performed as described by Wu et al. (2006). Briefly, total RNA was extracted from seven to 10 replicated biological samples using TRIZOL reagent (Invitrogen, http://www.invitrogen.com) following the manufacturer's instructions. The cDNA synthesis was carried out using a first-strand cDNA synthesis kit (Invitrogen, http://www.invitrogen.com); 1 μg of total RNA from No samples was subjected to reverse transcription using oligo(dT) and Superscript II reverse transcriptase (Invitrogen, http://www.invitrogen.com). The obtained cDNA was used as a template to generate WIPK, LOX3, NPR1, and JAR4 DNA fragments using PCR primers designed on Na’s published sequences. The PCR amplification was done in a final volume of 50 μL containing 10 ng of cDNA, 13 μL PCR buffer, 1.5 mm MgCl2, 0.2 mm of each primer, 200 mm of each dNTP, and 1.25 units of Taq DNA polymerase. The PCR conditions were as follows: an initial denaturation step at 94°C for 5 min, 30 cycles at 94°C for 1 min, 57°C for 1 min (50°C during the first two cycles), 72°C for 2 min, and a final extension step at 72°C for 10 min. The PCR fragments were gel purified and cloned into pGEM-T Easy vectors (Promega, http://www.promega.com) and then sequenced. Sequencing was performed using an ABI PRISM 377 automated DNA sequencer (Global Medical Instrumentation, http://www.gmi-inc.com). Each clone was sequenced in both sense and antisense directions and at least four clones were sequenced for each fragment. All the sequences have been deposited in the GenBank database under the following accession numbers: HM362911 (WIPK), HM362912 (LOX3), HM362910 (NPR1), HM362913 (JAR4), and HM362914 (Actin). TPI sequence was published by Wu et al. (2006) under the following accession: DQ158201.
SYBR Green Real-Time PCR Assay (Quantitative PCR)
Quantitative PCR (q-PCR) analysis was conducted using four to seven replicated biological samples for each time point in the kinetic analysis. The first step of reverse transcription was optimized to minimize errors that can be generated during cDNA synthesis; therefore, all total RNA samples were diluted to 0.5 mg/mL in 96-well PCR plates and the same enzyme master mix reaction was used for all samples. Two microliters of each diluted RNA sample was reverse transcribed as described in the previous section; the obtained cDNA samples were further diluted with water to 40 μL. q-PCR was performed on an ABI PRISM 7700 sequence detection system (Applied Biosystems, http://www.appliedbiosystems.com) using qPCR Core kits (Eurogentec, http://www.eurogentec.com). For each analysis, a linear standard curve, threshold cycle number versus log (designated transcript level) was constructed using serial dilutions of a specific cDNA standard; the levels of the transcript in all unknown samples were determined according to the standard curve. Actin, a housekeeping gene from Na and No that has been shown to have constant transcript levels by both RNA gel blotting and q-PCR after W + OS treatments (J. Wu, unpublished data) was used as an internal standard for normalizing cDNA concentration variations. The primers for the SYBR Green-based q-PCR were specifically designed to amplify in N×o transcripts from only Na or No, but not both. A PCR test was performed for each pair of primers and the product was visualized on agarose gel (Supplemental Fig. S6). In contrast, actin primers were designed to amplify both Na and No transcripts. Primer sequences were designed following the PCR conditions recommended by the manufacturer.
Statistical Analysis
Transcript and metabolic data, obtained at specific time points per treatment (wounding, OS elicitations, and MeJA), were analyzed with unpaired t tests using StatView statistical software (SAS Institute). We considered as significant only differences in transcript levels that are correlated with significant variations in metabolite levels. In other transcripts, e.g WIPK and NPR1, only comparisons that show more than 2-fold differences are considered as significant.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers HM362910 to HM362914.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. OS elicitation enhances the accumulation of uniparental transcript patterns of WIPK and LOX3, but not NPR1, in N×o lines (2–5).
Supplemental Figure S2. Both cis- and trans-regulatory elements act on the expression of WIPK, LOX3, and NPR1 genes elicited by M. sexta OS.
Supplemental Figure S3. Patterns of OS-elicited SA and JA accumulation in N×o 2 to 5 after OS elicitation.
Supplemental Figure S4. N×o lines (2–5) enhance the accumulation of only one parental transcript of JAR4 and TPI, and accumulated different levels of JA-Ile and TPI activity after M. sexta OS elicitation.
Supplemental Figure S5. MeJA treatment of N×o 1 to 5 elicits transcripts of both parental LOX3, JAR4, and TPIs, demonstrating that the uniparental pattern of transcript accumulation observed after OS elicitation is not due to gene inactivation.
Supplemental Figure S6. PCR products amplified in Na and No cDNA using reverse transcription primer pairs specific for Na and No WIPK, LOX3, JAR4, TPI, and NPR1.
Supplementary Material
Acknowledgments
We thank Siham Bezzi, Dr. Klaus Gase, and Dr. Tamara Krügel for invaluable technical support.
References
- Adams KL, Percifield R, Wendel JF. (2004) Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics 168: 2217–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams KL, Wendel JF. (2005) Polyploidy and genome evolution in plants. Curr Opin Plant Biol 8: 135–141 [DOI] [PubMed] [Google Scholar]
- Anssour S, Krugel T, Sharbel TF, Saluz HP, Bonaventure G, Baldwin IT. (2009) Phenotypic, genetic and genomic consequences of natural and synthetic polyploidization of Nicotiana attenuata and Nicotiana obtusifolia. Ann Bot (Lond) 103: 1207–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Baldwin IT, Staszak-Kozinski L, Davidson R. (1994) Up in smoke: I. Smoke-derived germination cues for post-fire annual, Nicotiana attenuata torr. Ex. Watson. J Chem Ecol 20: 2345–2371 [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. (2004) Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase MW, Knapp S, Cox AV, Clarkson JJ, Butsko Y, Joseph J, Savolainen V, Parokonny AS. (2003) Molecular systematics, GISH and the origin of hybrid taxa in Nicotiana (Solanaceae). Ann Bot (Lond) 92: 107–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ. (2007) Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol 58: 377–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson JJ, Knapp S, Garcia VF, Olmstead RG, Leitch AR, Chase MW. (2004) Phylogenetic relationships in Nicotiana (Solanaceae) inferred from multiple plastid DNA regions. Mol Phylogenet Evol 33: 75–90 [DOI] [PubMed] [Google Scholar]
- Comai L, Tyagi AP, Winter K, Holmes-Davis R, Reynolds SH, Stevens Y, Byers B. (2000) Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 12: 1551–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Meaux J, Pop A, Mitchell-Olds T. (2006) Cis-regulatory evolution of chalcone-synthase expression in the genus Arabidopsis. Genetics 174: 2181–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diezel C, von Dahl CC, Gaquerel E, Baldwin IT. (2009) Different lepidopteran elicitors account for cross-talk in herbivory-induced phytohormone signaling. Plant Physiol 150: 1576–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Liu B, Segal G, Abbo S, Levy AA, Vega JM. (1997) Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics 147: 1381–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L, Udall J, Nettleton D, Wendel J. (2008) Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol 6: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta RT, Pires JC, Iniguez-Luy F, Leon E, Osborn TC. (2007) Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19: 3403–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Doebley JF. (1997) DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci USA 94: 6809–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han FP, Fedak G, Ouellet T, Liu B. (2003) Rapid genomic changes in interspecific and intergeneric hybrids and allopolyploids of Triticeae. Genome 46: 716–723 [DOI] [PubMed] [Google Scholar]
- Horn M, Patankar AG, Zavala JA, Wu J, Doleckova-Maresova L, Vujtechova M, Mares M, Baldwin IT. (2005) Differential elicitation of two processing proteases controls the processing pattern of the ptrypsin proteinase inhibitor precursor in Nicotiana attenuata. Plant Physiol 139: 375–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang CX, Wright RJ, El-Zik KM, Paterson AH. (1998) Polyploid formation created unique avenues for response to selection in Gossypium (cotton). Proc Natl Acad Sci USA 95: 4419–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JH, Baldwin IT. (2006) Isolation and characterization of the threonine deaminase promoter in Nicotiana attenuata. Plant Sci 171: 435–440 [DOI] [PubMed] [Google Scholar]
- Kenton A, Parokonny AS, Gleba YY, Bennett MD. (1993) Characterization of the Nicotiana tabacum genome by molecular cytogenetics. Mol Gen Genet 240: 159–169 [DOI] [PubMed] [Google Scholar]
- Kitamura S, Inoue M, Ohmido N, Fukui K. (1997) Identification of parental chromosomes in the interspecific hybrids of Nicotiana rustica × N. tabacum and N. gosseidomin × N. tabacum, using genomic in situ hybridization. Breed Sci 47: 67–70 [Google Scholar]
- Kotseruba V, Gernand D, Meister A, Houben A. (2003) Uniparental loss of ribosomal DNA in the allotetraploid grass Zingeria trichopoda (2n = 8). Genome 46: 156–163 [DOI] [PubMed] [Google Scholar]
- Krügel T, Lim M, Gase K, Halitschke R, Baldwin IT. (2002) Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology 12: 177–183 [Google Scholar]
- Lagercrantz U, Lydiate DJ. (1996) Comparative genome mapping in Brassica. Genetics 144: 1903–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch IJ, Hanson L, Lim KY, Kovarik A, Chase MW, Clarkson JJ, Leitch AR. (2008) The ups and downs of genome size evolution in polyploid species of Nicotiana (Solanaceae). Ann Bot (Lond) 101: 805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KY, Matyasek R, Kovarik A, Leitch AR. (2004) Genome evolution in allotetraploid Nicotiana. Biol J Linn Soc Lond 82: 599–606 [Google Scholar]
- Lim KY, Soltis DE, Soltis PS, Tate J, Matyasek R, Srubarova H, Kovarik A, Pires JC, Xiong Z, Leitch AR. (2008) Rapid chromosome evolution in recently formed polyploids in Tragopogon (Asteraceae). PLoS ONE 3: e3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KY, Souckova-Skalicka K, Sarasan V, Clarkson JJ, Chase MW, Kovarik A, Leitch AR. (2006) A genetic appraisal of a new synthetic Nicotiana tabacum (Solanaceae) and the kostoff synthetic tobacco. Am J Bot 93: 875–883 [DOI] [PubMed] [Google Scholar]
- Lou Y, Baldwin IT. (2003) Manduca sexta recognition and resistance among allopolyploid Nicotiana host plants. Proc Natl Acad Sci USA 100: 14581–14586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens LN, Pires JC, Leon E, Vogelzang R, Oslach L, Osborn T. (2006) Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol 140: 336–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP, Whitton J. (2000) Polyploid incidence and evolution. Annu Rev Genet 34: 401–437 [DOI] [PubMed] [Google Scholar]
- Pearse IS, Krugel T, Baldwin IT. (2006) Innovation in anti-herbivore defense systems during neopolypoloidy—the functional consequences of instantaneous speciation. Plant J 47: 196–210 [DOI] [PubMed] [Google Scholar]
- Pontes O, Neves N, Silva M, Lewis MS, Madlung A, Comai L, Viegas W, Pikaard CS. (2004) Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc Natl Acad Sci USA 101: 18240–18245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayapuram C, Baldwin IT. (2007) Increased SA in NPR1-silenced plants antagonizes JA and JA-dependent direct and indirect defenses in herbivore-attacked Nicotiana attenuata in nature. Plant J 52: 700–715 [DOI] [PubMed] [Google Scholar]
- Sembdner G, Atzorn R, Schneider G. (1994) Plant hormone conjugation. Plant Mol Biol 26: 1459–1481 [DOI] [PubMed] [Google Scholar]
- Sembdner G, Parthier B. (1993) The biochemistry and the physiological and molecular actions of jasmonates. Annu Rev Plant Physiol Plant Mol Biol 44: 569–589 [Google Scholar]
- Shaked H, Kashkush K, Ozkan H, Feldman M, Levy AA. (2001) Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13: 1749–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker RC, Polzin K, Labate J, Specht J, Brummer EC, Olson T, Young N, Concibido V, Wilcox J, Tamulonis JP, et al. (1996) Genome duplication in soybean (Glycine subgenus soja). Genetics 144: 329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalicka K, Lim KY, Matyasek R, Matzke M, Leitch AR, Kovarik A. (2005) Preferential elimination of repeated DNA sequences from the paternal, Nicotiana tomentosiformis genome donor of a synthetic allotetraploid tobacco. New Phytol 166: 291–303 [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. (2000) The role of genetic and genomic attributes in the success of polyploids. Proc Natl Acad Sci USA 97: 7051–7057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Lu P, Tang K, Osborn TC. (1995) Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc Natl Acad Sci USA 92: 7719–7723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I. (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16: 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I, Rowe ML. (2002) Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14: 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar RM, Springer NM. (2006) Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics 173: 2199–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JA, Ni Z, Scheen A-C, Koh J, Gilbert CA, Lefkowitz D, Chen ZJ, Soltis PS, Soltis DE. (2006) Evolution and expression of homeologous loci in Tragopogon miscellus (Asteraceae), a recent and reciprocally formed allopolyploid. Genetics 173: 1599–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dam NM, Horn M, Mare M, Baldwin IT. (2001) Ontogeny constrains systemic protease inhibitor response in Nicotiana attenuata. J Chem Ecol 27: 547–568 [DOI] [PubMed] [Google Scholar]
- Wang J, Tian L, Lee HS, Wei NE, Jiang H, Watson B, Madlung A, Osborn TC, Doerge RW, Comai L, et al. (2006) Genome wide non-additive gene regulation in Arabidopsis allotetraploids. Genetics 172: 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tian L, Madlung A, Lee HS, Chen M, Lee JJ, Watson B, Kagochi T, Comai L, Chen ZJ. (2004) Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics 167: 1961–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Halitschke R, Kang JH, Berg A, Harnisch F, Baldwin IT. (2007) Independently silencing two JAR family members impairs levels of trypsin proteinase inhibitors but not nicotine. Planta 226: 159–167 [DOI] [PubMed] [Google Scholar]
- Wendel JF. (2000) Genome evolution in polyploids. Plant Mol Biol 42: 225–249 [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. (2004) Evolutionary changes in cis and trans-gene regulation. Nature 430: 85–88 [DOI] [PubMed] [Google Scholar]
- Wu J, Baldwin IT. (2009) Herbivory-induced signaling in plants: perception and action. Plant Cell Environ 32: 1161–1174 [DOI] [PubMed] [Google Scholar]
- Wu J, Hettenhausen C, Baldwin I. (2006) Evolution of proteinase inhibitor defenses in north American allopolyploid species of Nicotiana. Planta 224: 750–760 [DOI] [PubMed] [Google Scholar]
- Wu J, Hettenhausen C, Meldau S, Baldwin IT. (2007) Herbivory rapidly activates MAPK signaling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. Plant Cell 19: 1096–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Borevitz JO. (2009) Global analysis of allele-specific expression in Arabidopsis thaliana. Genetics 182: 943–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.