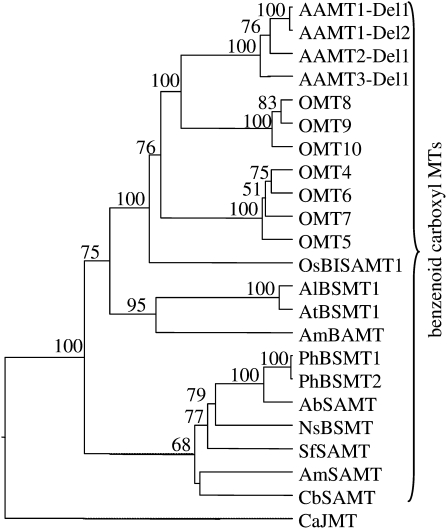
Figure 5.
Dendrogram analysis of maize AAMT1-Del1, AAMT1-Del2, AAMT2-Del1, AAMT3-Del1, OMT4, OMT5, OMT6, OMT7, OMT8, OMT9, and OMT10 with different methyltransferases (MTs) specific for carboxyl groups of benzenoids and jasmonic acid. The analysis was conducted using a neighbor-joining algorithm. Bootstrap values are shown in percentage and were generated with a sample of 1,000. Accession numbers are as follows: PhBSMT1, AAO45012; PhBSMT2, AAO45013; AbSAMT, BAB39396; NsBSMT, CAF31508; SfSAMT, CAC33768; AmSAMT, AAN40745; CbSAMT, AAF00108; AlBSMT1, AAP57211; AtBSMT1, NP_187755; AmBAMT, AAF98284; CaJMT, ABB02661; OsBISAMT1, AAS18419. Sequences of OMT4, OMT5, OMT6, OMT7, OMT9, and OMT10 were obtained from the Maize Genome Database (www.maizesequence.org) with the following accession numbers: OMT4, GRMZM2G143871; OMT5, GRMZM2G133996; OMT6, GRMZM2G050307; OMT7, GRMZM2G050321; OMT9, GRMZM2G303419; OMT10, GRMZM2G405947. The ORFs of OMT9 and OMT10 were reconstructed in silico by the deletion of a stop codon (OMT9) or the deletion of two frame-shift mutations (OMT10).