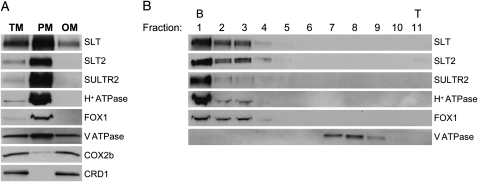
Figure 6.
Localization of the Chlamydomonas SLT1, SLT2, and SULTR2 transport proteins. A, Chlamydomonas strain D66 was grown in TAP medium to midlogarithmic phase and then grown in TAP−S for an additional 24 h. Total membranes (TM) were isolated and partitioned into plasma membrane fraction (polyethylene glycol fraction; PM) and a fraction containing all other membranes (dextran fraction; OM). A total of 20 μg of protein from each fraction was resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane for immunoblot analyses using antibodies against SLT, SLT2, SULTR2, plasma membrane H+-ATPase, FOX1 ferroxidase (plasma membrane), CRD1 (thylakoid membrane and chloroplast envelope), COX2b (mitochondrial inner membrane), and V-ATPase (tonoplast). B, To separate the plasma membrane from the tonoplast, 600 μg of protein from the polyethylene glycol phase was loaded onto a 15% to 45% Suc gradient and separated into 11 fractions of equal volume (fraction 1 is at the bottom [B] of the gradient and fraction 11 is at the top [T] of the gradient). The proteins of each fraction were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane for immunoblotting, as described in “Materials and Methods.”