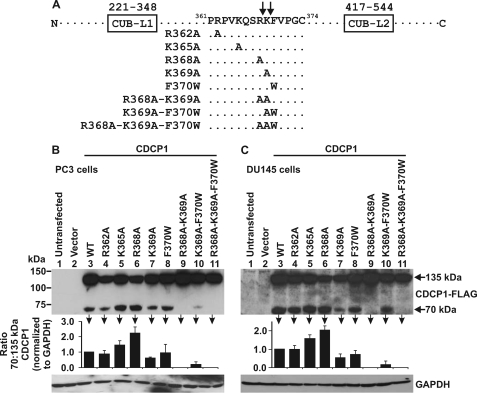
FIGURE 4.
Examination of the cellular proteolytic processing sites of CDCP1. A, schematic showing the location of single, double, and triple amino acid mutations introduced into the CDCP1-Flag sequence between CUB-like domain 1 and 2 (CUB-L1 and CUB-L2), R362A, K365A, R368A, K369A, F370W, R368A-K369A, K369A-F370W, and R368A-K369A-F370W. B, PC3 and C, DU145 cell lysates from cells either untransfected or transiently transfected with either vector or wild type or mutant CDCP1-Flag expression constructs analyzed by anti-Flag Western blot analysis. Anti-GAPDH Western blot analysis was performed to examine protein loading. Graphs were generated from densitometric analysis of at least three separate experiments. For 70 and 135 kDa CDCP1 and GAPDH, signal intensities from cells transfected with mutant constructs were normalized against the signal from cells transfected with wild-type CDCP1. These normalized values were used to calculate the ratio of 70 to 135 kDa CDCP1.