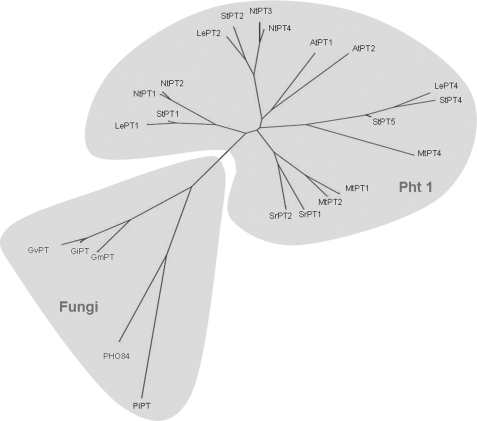
FIGURE 1.
Unrooted phylogenetic relationship of PiPT with other high affinity phosphate transporters from plants and fungi. Protein names are followed by GenBankTM (GB) accession numbers: GmPT (DQ074452) from Glomus mosseae; GiPT (AF359112) from Glomus intraradices; GvPT (U38650) from Glomus verisforme; PHO84 (D90346) from S. cerevisiae; LePT1 (AF022873), LePT2 (AF022874), and LePT4 (AY885651) from Lycopersicon esculentum; AtPT1 (U62330) and AtPT2 (U62331) from A. thaliana; StPT1 (X98890), StPT2 (X98891), StPT4 (AY793559), and StPT5 (AY885654) from Solanum tuberosum; MtPT1 (AF000354), MtPT2 (AF000355), and MtPT4 (AY116210) from Medicago truncatula; SrPT1 (AJ286743) and SrPT2 (AJ286744) from Sesbania rostrata; NtPT1 (AF156696), NtPT2 (AB042950), NtPT3 (AB042951), and NtPT4 (AB042956) from Nicotiana tobacum. The evolutionary history was inferred using the Neighbor-Joining method (72). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method (73) and are in the units of the number of base substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (Complete Deletion option). Phylogenetic analyses were done by using MEGA4 (74).