Abstract
Selenophosphate synthetase 1 (SPS1) is an essential cellular gene in higher eukaryotes. Five alternative splice variants of human SPS1 (major type, ΔE2, ΔE8, +E9, +E9a) were identified wherein +E9 and +E9a make the same protein. The major type was localized in both the nuclear and plasma membranes, and the others in the cytoplasm. All variants form homodimers, and in addition, the major type forms a heterodimer with ΔE2, and ΔE8 with +E9. The level of expression of each splice variant was different in various cell lines. The expression of each alternative splice variant was regulated during the cell cycle. The levels of the major type and ΔE8 were gradually increased until G2/M phase and then gradually decreased. ΔE2 expression peaked at mid-S phase and then gradually decreased. However, +E9/+E9a expression decreased gradually after cell cycle arrest. The possible involvement of SPS1 splice variants in cell cycle regulation is discussed.
Keywords: alternative splicing, cell cycle, selenium, selenocysteine, selenophosphate synthetase 1
1. Introduction
Selenium is an essential trace element and provides many health benefits for animals including humans. For example, this element has been known to play a role in cancer prevention, aging retardation, immune augmentation, prevention of cardiovascular disease, muscle development and immune function [1-4]. Most of these various selenium effects are believed to be mediated by selenoproteins which contain the selenium-containing amino acid, selenocysteine (Sec), at their active site(s). Sec, which is the 21st amino acid in the genetic code, is inserted into a growing peptide during translation in response to the codon, UGA [5-7]. The biosynthesis of Sec is carried out through a unique pathway that is distinct from that of other amino acids. Most amino acids are synthesized prior to being aminoacylated onto their cognate tRNAs; however, Sec is synthesized on its tRNA, designated tRNA[Ser]Sec. In eukaryotes, tRNA[Ser]Sec is initially aminoacylated with serine by seryl-tRNA synthetase, the hydroxyl moiety of serine on seryl-tRNA[Ser]Sec is then replaced by phosphate group to form O-phosphoseryl-tRNA[Ser]Sec that is catalyzed by a specific kinase, phosphoseryl-tRNA[Ser]Sec kinase [8]. Finally, the phosphate group on O-phosphoseryl-tRNA[Ser]Sec is exchanged with activated selenium to form selenocysteyl-tRNA[Ser]Sec catalyzed by the enzyme, Sec synthase (SecS) [9,10]. The O-phosphoseryl-tRNA[Ser]Sec intermediate is not synthesized in eubacteria wherein SecS in E. coli (SelA) removes the hydroxyl group of serine on seryl-tRNA[Ser]Sec to form aminoacrylyl-tRNA[Ser]Sec and the resulting aminoacrylyl intermediate in turn accepts activated selenium to form Sec-tRNA[Ser]Sec [11]. Mono-selenophosphate (SeP) is the activated selenium form in both bacteria and eukaryotes [12], and it is synthesized from selenide and ATP. Selenophosphate synthetase (SPS) catalyzes the synthesis of SeP [13]. Two isoforms of SPS were found in higher eukaryotes, SPS1 and SPS2, whereas only one type of SPS (SelD) exists in lower eukaryotes and eubacteria [14]. The sequence homology between SPS1 and SPS2 is different depending on the species. For example, the amino acid sequence homology between human SPS1 and SPS2 is 72% and that between Drosophila SPS1 and SPS2 is approximately 45%. One of the major differences between SPS1 and SPS2 is that SPS1 has an arginine at the position corresponding to Sec in SPS2 [15,16]. The sequence homology between Drosophila SPS1 and Drosophila SPS2 is 45%, while that between human SPS1 and Drosophila SPS1 is 71%, indicating that the SPS1 sequence has been well conserved during evolution (NCBI, Build 37.1 for human sequence and BDGP Release 5.12 for Drosophila).
Recently, it was shown that SPS1 does not have SeP synthesis activity, but SPS2 catalyzes SeP synthesis. In in vitro experiments, SPS2 synthesized SeP from selenide and ATP, but SPS1 did not have this activity [9]. The ablation of SPS2 mRNA in NIH3T3 cells led to the loss of selenoprotein biosynthesis, while the inhibition of SPS1 expression did not affect the biosynthesis of selenoprotein, and the synthesis of selenoproteins in the cells in which SPS2 was knocked down was rescued by expression of SPS2, but not by SPS1 [17]. Although some insects such as the red beetle and silkworm have lost the selenoprotein synthesizing machinery including SPS2, SPS1 is still encoded in the genome of those organisms suggesting SPS1 is not essential for selenoprotein synthesis [18].
Although SPS1 does not catalyze SeP biosynthesis [12], there are several lines of evidence suggesting that it plays important roles in the cell. Targeted removal of the gene encoding SPS1 (also designated patufet) in Drosophila causes embryonic lethality suggesting that SPS1 participates in pathways important for cell viability [19]. It was also found that knockout of SPS1 caused an increase in reactive oxygen species (ROS) and the haploinsufficiency of SPS1 suppressed the abnormal phenotypes caused by overexpression of genes involved in Ras-regulated signaling pathway [20,21]. These latter findings suggest that SPS1 participates in the regulation of oxidative stress and cell growth. In agreement with this predicted function of SPS1, knockdown of Drosophila SPS1 led to growth inhibition and ROS generation [22]. Interestingly, knockdown of Drosophila SPS1 caused generation of megamitochondria through the increase of intracellular glutamine levels.
When cDNA of SPS1 was cloned from human lung adenocarcinoma and transformed into a SelD deficient mutant of E. coli, the selD mutation was not complemented in selenite containing medium, but complemented when the cells were cultured in the medium supplemented with L-Sec [23]. From these findings, it was suggested that SPS1 is involved in the recycling of Sec. However, the mechanism of Sec recycling and how SPS1 regulates Sec recycling have not been determined. Interestingly, SPS1 was found to interact with soluble liver antigen (SLA) which was recently identified as eukaryotic Sec synthase (SecS) and the binding reaction was enhanced by a protein designated SECp43 [24].
In the present study, we found that hSPS1 produces five alternative splice variants and one of them was generated by mis-splicing. These alternative splice variants showed unique characteristics of intermolecular interaction, subcellular localization, specific tissue and cell cycle expression suggesting that each splice variant has a specific function for cell growth.
2. Materials and methods
2.1. Cell culture and DNA transfection
A549, CRL7407, MCF7, Chang liver, HeLa, and 293 cells were grown in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum (FBS) (Hyclone) and 1% antibiotic-antimycotic (AA), and BJAB cells in RPMI 1640 medium supplemented with 10% FBS and 1% AA. All cultures were maintained in a humidified atmosphere of 5% CO2 at 37°C. Transient transfections in HeLa cells were carried out using Lipofectamine Plus (Invitrogen) according to the manufacturer’s instructions and calcium phosphate method was used in 293 cell transfection.
2.2. Vector constructions
Vectors that express alternative splice variants were constructed in two steps: In the first step, a pool of clones containing all types of splice variants was prepared. In the second step, each splice variant was screened using colony PCR. Vectors that can express Flag- or HA-tagged proteins were prepared by PCR using primers that contain the sequences encoding Flag or HA, respectively (see Supplementary Materials and methods for detail).
2.3. Immunostaining and confocal microscopy
HeLa cells transfected with Flag-tagged hSPS1 expression vector were transferred onto a cover glass one day before staining. Immunocytochemistry was carried out as described previously [22] with a slight modification. After the cells were fixed, permeabilized and blocked, anti-Flag antibody (Sigma) diluted in 3% BSA in PBS (1:2,500) was treated for 1 h at room temperature. The cells were then incubated with Alexa 488 anti-mouse IgG (Invitrogen) diluted in 3% BSA in PBS (1:2,000) for 1 h at room temperature. For counterstaining, cells were stained with 50 μg/ml of propidium iodide (PI) (Sigma) for 20 min at room temperature. Cells were observed with a LSM 510 NLO confocal microscope (Carl Zeiss). Excitation was at 488 nm for Alexa 488 and 543 nm for PI.
2.4. Quantitative real-time RT-PCR
To determine the number of hSPS1 mRNA molecules, total RNA was isolated from cells using TRIzol reagent (Invitrogen), first-strand cDNA was synthesized from 3 μg of total RNA with Mo-MuLV reverse transcriptase (Super-Bio), and used for the PCR reaction. cDNAs were amplified with SYBR Green PCR master mix and specific primers using the ABI 7300 real-time PCR system (Applied Biosystems). Vectors which can express the hSPS1 alternative splice variant were serially diluted from 101 to 108 copy numbers and applied to real-time PCR. Standard curve was obtained by plotting the log of the number of DNA templates versus the measured CT (cycle-threshold). Using the equation calculated from the standard curve, the copy number of hSPS1 mRNA per nanogram of total RNA was determined.
2.5. Immonoprecipitation and immunoblotting
A vector which can express a Flag-tagged splice variant was cotransfected with a HA-tagged splice variant expression vector in 293 cells in a manner that a vector expressing one type of Flag-tagged splice variant was cotransfected with a vector expresses one of all types of HA-tagged splice variants. Cell lysates were coimmunoprecipitated with anti-Flag M2 affinity gel (Sigma) and the precipitates subjected to western blotting using anti-HA antibody or anti-Flag antibody (see Reference [22] and Supplementary Materials and methods for detail).
2.6. Synchronization of cells and flow cytometry
For synchronization, HeLa cells were incubated with 5 μg/ml of aphidicolin (Sigma) for 24 h. Cells were washed with fresh medium and released from the aphidicolin block for indicated times. Cell cycle profiles were analyzed by flow cytometry with standard PI staining methods. Asynchronized and synchronized cells were harvested and washed once with PBS. Cells were fixed overnight in 70% ethanol at -20°C, washed with PBS twice and then resuspended with PBS containing 200 μg/ml of RNase A. After 10 min incubation at room temperature, cells were further incubated with 100 μg/ml of PI in PBS for 30 min at room temperature. Stained cells were analyzed by using a FACS Calibur flow cytometer and CellQuest Pro software (BD Biosciences).
3. Results
3.1. Identification of alternative splice variants in hSPS1 mRNA
To elucidate the molecular function of hSPS1, its cDNA was amplified using total cDNAs prepared from HeLa cells and cloned into a vector. The sequences of several clones harboring hSPS1 cDNA were determined. Interestingly, we found that one of the clones was a truncated form in which exon 8 was deleted. We, therefore, assumed that there would be alternative splice variants in hSPS1 mRNA. To find possible alternative splice forms, we searched EST clones from the University of California, Santa Cruz Genome Browser (http://genome.ucsc.edu) in scrutiny. Among a total of 283 EST clones of hSPS1, 277 ESTs were the same as known hSPS1 mRNA and designated major type (MT). Except MT, there were two ESTs (CX787008, DB001684) in which exon 2 was deleted (ΔE2), two ESTs (AI741523, AI742171) where exon 8 was deleted (ΔE8) and additional 2 ESTs (BX385308, CR604492) where exon 9 was added to the major type (+E9). These data suggest that there are 4 alternative splice variants of hSPS1 mRNA in human cells.
To confirm that the predicted alternative splice variants are present in human cells, we performed RT-PCR in several steps and surprisingly, detected 5 splice variants instead of the 4 predicted splice variants (see Supplementary Result and Supplementary Fig. S1). The sequence of each splice variant was determined and found to be consistent with the expected sequence (GenBank Accession No. GU954545, GU954546, GU954547, GU954548, and GU954549; Supplementary Fig. S2).
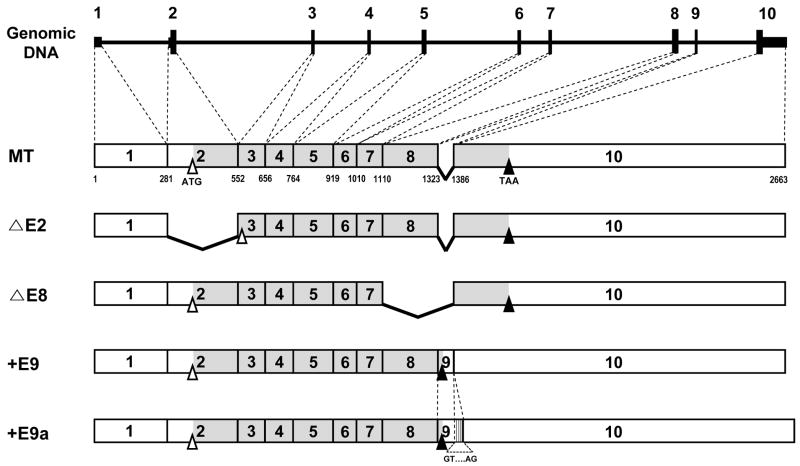
As summarized in Fig. 1, hSPS1 pre-mRNA is alternatively spliced to produce 5 different splice variants. The translation initiation codon is located in exon 2 and the termination codon in exon 10. As expected, ΔE2 and ΔE8 do not have exon 2 and exon 8, respectively. Since the normal initiation codon is located in exon 2, ΔE2 cannot use the original initiation codon. Therefore, ΔE2 must use another initiation codon. In exon 3 (at +561), there is an ATG codon and the surrounding Kozak’s consensus sequence. Both forms are void of exon 9 as found in MT. The sequence of exon 9 is very short and there is the translation termination codon (TAA) within exon 9 suggesting that the translated protein from +E9 will be truncated from MT, even though the size of mRNA of +E9 is larger than that of MT. +E9a has an additional sequence which is part of 5’ region of intron 9. Interestingly, the beginning and end of the additional sequence contain splice junction sequences (GT and AG), respectively. In the genomic DNA sequence, there is no gap between exon 9 and the additional sequence. This suggests that +E9a is generated by mis-splicing. The protein sequences of +E9 and +E9a should be the same, because there is a termination codon within exon 9.
Fig. 1. Five types of alternative splice variants of hSPS1 mRNA.
The black line on the top represents genomic DNA, on which exons are shown as a bar. Numbers designate exons of each predicted alternative splice variant. The open and filled area in the exons designate the untranslated region and the open reading frame, while open and closed triangles designate the position of translational initiation and termination codons, respectively. Bent lines within cDNA designate a deleted exon in each splice variant. The numbers underneath MT indicate nucleotide positions.
3.2. Subcellular localizations of splice variants
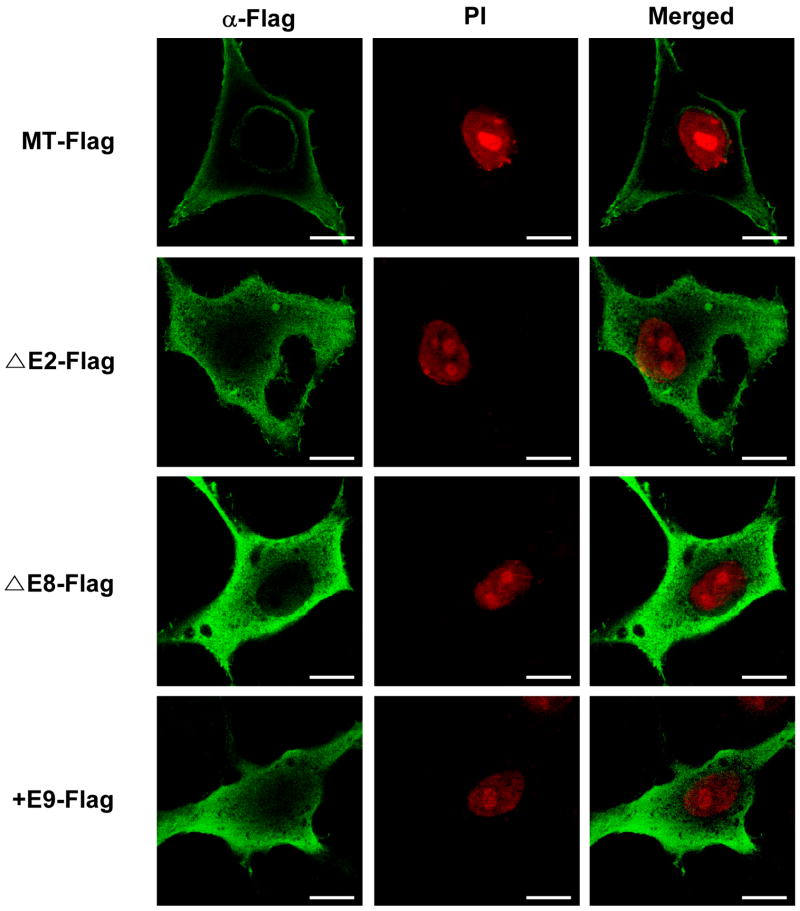
Recently, it was reported that hSPS1 makes supramolecular complexes by interacting with SecS both in vivo and in vitro and this supramolecular complex shuttles between the nucleus and the cytoplasm [24]. Therefore, it is important to determine the subcellular localization of each alternative splice variant to elucidate their functions. The Flag-tagged splice variants were expressed and detected by immunocytochemistry using anti-Flag antibody. As shown in Fig. 2, MT was localized on both plasma and the nuclear membrane, but the other three types were localized in the cytoplasm homogeneously. These results suggest that each alternative splice type plays a unique role by interacting with different proteins or molecules.
Fig. 2. Subcellular localization of hSPS1 alternative splice variants.
Flag-tagged hSPS1 overexpression vectors which can express each alternative splice variant were transfected into HeLa cells. Forty eight hours after transfection, cells were fixed and permeabilized, then hSPS1 was detected by mouse anti-Flag antibody and Alexa 488 anti-mouse IgG (green). Nucleus was stained with PI (red) and cells were observed under the confocal microscope. Scale bars represent 10 μm.
3.3. Interactions between splice variants
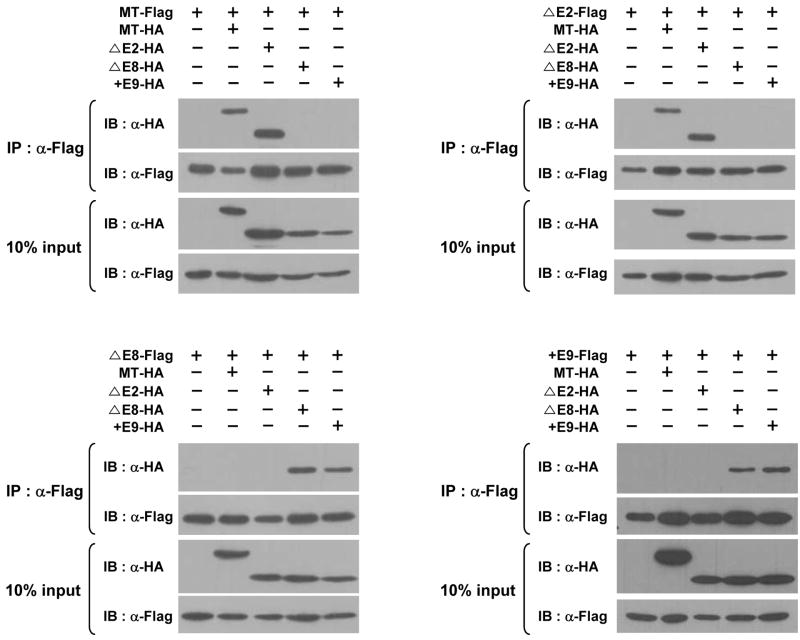
By analyzing the X-ray crystallography structure of hSPS1, it was suggested recently that hSPS1 could form a homodimer [25]. We therefore examined whether all the splice variants could make dimers (homodimers as well as heterodimers). In addition to Flag-tagged constructs, HA-tagged constructs were also prepared (see Supplementary Materials and methods). After co-transfecting a Flag-tagged construct and a HA-tagged construct into 293 cells, cell lysates were subjected to immunoprecipitation using anti-Flag antibody and then the co-purified products were detected by western blot analysis using anti-Flag or anti-HA antibody. As shown in Fig. 3, MT-Flag interacts with MT-HA and ΔE2-HA, and ΔE2-Flag with MT-HA and ΔE2-HA. Similarly, ΔE8-Flag interacts with ΔE8-HA and +E9-HA and +E9-Flag with ΔE8-HA and +E9-HA. These results suggest that all the splice variants can make homodimers as well as heterodimers between MT and ΔE2, and between ΔE2 and +E9. Although it cannot be excluded that they form multimers such as trimers or tetramers, each splice variant makes specific interactions. It should be noted that MT forms a heterodimer only with ΔE2, and the C-terminal sequences of them are the same suggesting C-terminal sequence is important for heterodimer formation.
Fig. 3. Interactions between each hSPS1 splice variant.
Flag or HA tagged splice variant expression vectors were transfected in 293 cells in the combinations shown, extracts were prepared, splice variants were immunoprecipitated, electrophoresed and detected by western blotting.
3.4. Expression of alternative splice variants during cell cycle progression
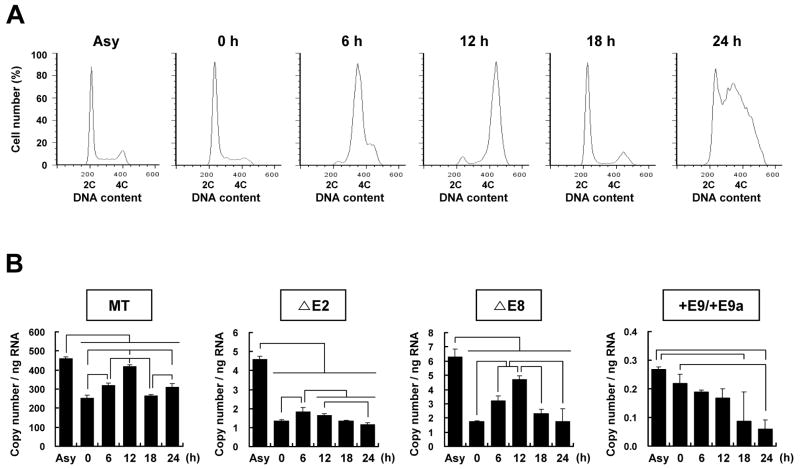
It has been reported that patufet which is the Drosophila homolog of SPS1 is expressed highly in actively dividing cells and its deletion caused embryonic lethality during fly development [19,26]. More recently it was also reported that knockdown of Drosophila SPS1 led to growth inhibition of cells [22]. These reports suggest that the expression of hSPS1 can be controlled during the cell cycle. We examined the expression level of alternative splice variants during the cell cycle. After synchronizing the cells at G1/S phase, the synchronized cells were then released to proceed through the cell cycle by removing aphidicolin. At 6, 12, and 18 h after release, cells were in the mid-S phase, in the G2/M phase, and in the G1/S phase, respectively. However, at 24 h after release, the cells were detected in all phases of the cell cycle (Fig. 4A).
Fig. 4. Expression of hSPS1 alternative splice variants during cell cycle progression.
(A) FACS (fluorescent associated cell sorter) analysis of cells after release from synchronization. X axis represents DNA contents and Y axis % of cell number. (B) Quantification of the amount of splice variants during the cell cycle. Real-time RT-PCR was performed using the same primers given in Supplementary Table S2. Experiments were performed in triplicate, and error bars denote the standard deviation from the mean of three experiments. Statistical significance was tested by pairwise Student’s t-test with Bonferroni correction for multiple testing (p<0.05). Asy represents asynchronized cells.
The expression levels of each alternative splice variant were measured by real-time PCR as described in the Materials and methods. As shown in Fig. 4B, the expression of the MT variant was gradually increased until the G2/M phase and then decreased. For example, cells arrested at the G1/S phase by aphidicolin treatment showed that the MT variant expression level was decreased to 56% compared to that in asynchronized cells (P<0.05). The level of MT expression in the cells of the G2/M phase was increased to 160% compared to that of cells arrested at the G1/S phase (P<0.05). The levels of MT were similar at 0 h and 18 h when the cells were in G1/S phase. ΔE2 expression was peaked at mid-S phase and then gradually decreased. ΔE8 showed similar expression patterns as MT, but +E9/+E9a levels were decreased gradually as time lapsed suggesting the formation of +E9/+E9a was impaired by synchronization. These results suggest that alternative splicing of hSPS1 mRNA is closely related with cell cycle control.
4. Discussion
Five alternative splice variants of hSPS1 mRNA (MT, ΔE2, ΔE8, +E9 and +E9a) were identified in HeLa cells. Through searching the web browser of the University of California at Santa Cruz, four of these variants could be predicted with only the exception of +E9a. Interestingly, +E9a contains an extra 29 nucleotide extension of +E9. In genomic DNA, there is no gap between the 3’ end of +E9 and the 5’ end of +E9a. Furthermore, the consensus sequence of intron/exon junction (GT/AG) occurs at the 5’ and 3’ boundary of the additional sequence of +E9a suggesting +E9a is generated by attaching a part of intron 9 to the exon 9 by mis-splicing (see Fig. 1 and Supplementary Fig. S2).
The average amount of each splice variant was different. Approximately 90% of SPS1 population consists of MT. ΔE2 and ΔE8 comprise 2% of the total SPS1 mRNA, while +E9 and +E9a make up less than 1%.
Although there are 5 alternative splice variants in hSPS1 mRNA, only four proteins are produced, because there is a translational termination codon in exon 9. In terms of protein size, the MT protein is the largest, ΔE2 the second, +E9 /+E9a the third and ΔE8 the smallest.
The hSPS1 splice variants can interact with specific combinations; MT with ΔE2 and ΔE8 with +E9. It should be noted that the C-terminal sequences of MT and ΔE2 are the same and the C-terminus of ΔE8 and +E9 are truncated. These results suggest that the C-terminus of SPS1 is important for interaction.
As described in the Introduction, SPS1 participates in Ras mediated cell signaling in Drosophila [20,21]. MT is located on both the plasma membrane and nuclear membrane and the other splice variants in the cytoplasm. All splice variants interact in a specific manner. Therefore, it can be speculated that the interactions of SPS1 between these molecules mediate cellular signal transductions. However, interactions of splice variants with other proteins have not been found. Because only MT is localized in the membranes, membrane localizing signals may occur in MT. However, we could not find any known common membrane localizing signals suggesting MT may interact with other membrane localizing protein(s).
Each splice variant was expressed differentially depending on the transformed cell lines derived from various tissues. For example, MT and +E9/+E9a are expressed in highest levels in BJAB cells which were derived from B cells; ΔE2 was expressed in the highest level in 293 cells which were derived from kidney; and ΔE8 was expressed in highest level in MCF7 cells which were derived from breast tissue. It is not clear whether these expression patterns are specific to each tissue or they are changed during transformation. However, if we compare Supplementary Fig. S3 with Supplementary Fig. S4, the expression pattern of splice variants at least in liver and kidney was changed during cell transformation. The expression level of splice variants is also regulated in a specific manner during the cell cycle and this result is consistent with the proposal that SPS1 is required for cell growth and that SPS1 is an essential gene; i.e., when Drosophila SPS1 was knocked out, development was halted at the embryonic stage. Our findings in the present study suggest that alternative splice variants of SPS1 play important roles in cell growth presumably by controlling cell cycle progression through their specific subcellular localization and interaction.
Supplementary Material
Acknowledgments
This work was supported by the Korea Research Foundation Grants funded by the Korean Government (MEST) (KRF-2005-070-C00086 and KRF-2008-005-J00201) to BJL, and in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. JYK, MSS, and KHL were supported by Brain Korea 21 Research Fellowship from the Korea Ministry of Education and Human Resources Development.
Abbreviations
- AA
antibiotic-antimycotic
- CT
cycle-threshold
- FACS
fluorescent associated cell sorter
- FBS
fetal bovine serum
- HA
haemagglutinin
- MT
major type
- PI
propidium iodide
- ROS
reactive oxygen species
- SPS
selenophosphate synthetase
- Sec
selenocysteine
- SecS
Sec synthase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hatfield DL, Berry MJ, Gladyshev VN. Selenium: Its molecular biology and role in human health. 2. Springer, NY: 2006. [Google Scholar]
- 2.Flohé L. Selenium in mammalian spermiogenesis. Biol Chem. 2007;388:987–995. doi: 10.1515/BC.2007.112. [DOI] [PubMed] [Google Scholar]
- 3.Papp LV, Lu J, Holmgren A, Khanna KK. From selenium to selenoproteins: synthesis, identity, and their role in human health. Antioxid Redox Signal. 2007;9:775–806. doi: 10.1089/ars.2007.1528. [DOI] [PubMed] [Google Scholar]
- 4.Boosalis MG. The role of selenium in chronic disease. Nutr Clin Pract. 2008;23:152–160. doi: 10.1177/0884533608314532. [DOI] [PubMed] [Google Scholar]
- 5.Lee BJ, Worland PJ, Davis JN, Stadtman TC, Hatfield DL. Identification of a selenocysteyl-tRNASer in mammalian cells that recognizes the nonsense codon, UGA. J Biol Chem. 1989;264:9724–9727. [PubMed] [Google Scholar]
- 6.Leinfelder W, Stadtman TC, Böck A. Occurrence in vivo of selenocysteyl-tRNASER in Escherichia coli. J Biol Chem. 1989;264:9720–9723. [PubMed] [Google Scholar]
- 7.Longtin R. A forgotten debate: Is selenocysteine the 21st amino acid? J Natl Cancer Inst. 2004;96:504–505. doi: 10.1093/jnci/96.7.504. [DOI] [PubMed] [Google Scholar]
- 8.Carlson BA, Xu XM, Kryukov GV, Rao M, Berry MJ, Gladyshev VN, Hatfield DL. Identification and characterization of phosphoseryl tRNA[Ser]Sec kinase. Proc Natl Acad Sci USA. 2004;101:12848–12853. doi: 10.1073/pnas.0402636101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu XM, Carlson BA, Mix H, Zhang Y, Saira K, Glass RS, Berry MJ, Gladyshev VN, Hatfield DL. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007;5:e4. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu XM, Carlson BA, Zhang Y, Mix H, Kryukov GV, Glass RS, Berry MJ, Gladyshev VN, Hatfield DN. New developments in selenium biochemistry: selenocysteine biosynthesis in eukaryotes and archaea. Biol Trace Elem Res. 2007;119:234–241. doi: 10.1007/s12011-007-8003-9. [DOI] [PubMed] [Google Scholar]
- 11.Forchhammer K, Böck A. Selenocysteine synthase from Escherichia coli. Analysis of the reaction sequence. J Biol Chem. 1991;266:6324–6328. [PubMed] [Google Scholar]
- 12.Glass RS, Singh WP, Jung W, Veres Z, Scholz TD, Stadtman TC. Monoselenophosphate: synthesis, chracterization, and identity with the prokaryotic biological selenium donor, compound SePX. Biochemistry. 1993;32:12555–12559. doi: 10.1021/bi00210a001. [DOI] [PubMed] [Google Scholar]
- 13.Ehrenreich A, Forchhammer K, Tormay P, Veprek B, Böck A. Purification and characterisation of the enzyme catalysing selenium activation. Eur J Biochem. 1992;206:767–773. doi: 10.1111/j.1432-1033.1992.tb16983.x. [DOI] [PubMed] [Google Scholar]
- 14.Leinfelder W, Forchhammer K, Veprek B, Zehelein E, Böck A. In vitro synthesis of selenocysteinyl-tRNAUCA from seryl-tRNAUCA: Involvement and characterization of the selD gene product. Proc Natl Acad Sci USA. 1990;87:543–547. doi: 10.1073/pnas.87.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Low SC, Harney JW, Berry MJ. Cloning and functional characterization of human selenophosphate synthetase, an essential component of selenoprotein synthesis. J Biol Chem. 1995;270:21659–21664. doi: 10.1074/jbc.270.37.21659. [DOI] [PubMed] [Google Scholar]
- 16.Guimaraes MJ, Peterson D, Vicari A, Cocks BG, Copeland NG, Gilbert DJ, Jenkins NA, Ferrick DA, Kastelein RA, Bazan JF, Zlotnik A. Identification of a novel selD homolog from Eukaryotes, Bacteria, and Archaea: Is there an autoregulatory mechanism in selenocysteine metabolism? Proc Natl Acad Sci USA. 1996;93:15086–15091. doi: 10.1073/pnas.93.26.15086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu XM, Carlson BA, Irons R, Mix H, Zhong N, Gladyshev VN, Hatfield DL. Selenophosphate synthetase 2 is essential for selenoprotein biosynthesis. Biochem J. 2007;404:115–120. doi: 10.1042/BJ20070165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lobanov AV, Hatfield DL, Gladyshev VN. Selenoproteinless animals: Selenophosphate synthetase SPS1 functions in a pathway unrelated to selenocysteine biosynthesis. Protein Sci. 2008;17:176–182. doi: 10.1110/ps.073261508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alsina B, Serras F, Baguñà J, Corominas M. patufet, the gene encoding the Drosophila melanogaster homologue of selenophosphate synthetase, is involved in imaginal disc morphogenesis. Mol Gen Genet. 1998;257:113–123. doi: 10.1007/s004380050630. [DOI] [PubMed] [Google Scholar]
- 20.Morey M, Serras F, Baguñà J, Hafen E, Corominas M. Modulation of the Ras/MAPK signalling pathway by the redox function of selenoproteins in Drosophila melanogaster. Dev Biol. 2001;238:145–156. doi: 10.1006/dbio.2001.0389. [DOI] [PubMed] [Google Scholar]
- 21.Morey M, Corominas M, Serras F. DIAP1 suppresses ROS-induced apoptosis caused by impairment of the selD/sps1 homolog in Drosophila. J Cell Sci. 2003;116:4597–4604. doi: 10.1242/jcs.00783. [DOI] [PubMed] [Google Scholar]
- 22.Shim MS, Kim JY, Jung HK, Lee KH, Xu XM, Carlson BA, Kim KW, Kim IY, Hatfield DL, Lee BJ. Elevation of glutamine level by selenophosphate synthetase 1 knockdown induces megamitochondrial formation in Drosophila cells. J Biol Chem. 2009;284:32881–32894. doi: 10.1074/jbc.M109.026492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura T, Yamamoto S, Takahata M, Sakaguchi H, Tanaka H, Stadtman TC, Inagaki K. Selenophosphate synthetase genes from lung adenocarcinoma cells: Sps1 for recycling L-selenocysteine and Sps2 for selenite assimilation. Proc Natl Acad Sci USA. 2004;101:16162–16167. doi: 10.1073/pnas.0406313101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Small-Howard A, Morozova N, Stoytcheva Z, Forry EP, Mansell JB, Harney JW, Carlson BA, Xu XM, Hatfield DL, Berry MJ. Supramolecular complexes mediate selenocysteine incorporation in vivo. Mol Cell Biol. 2006;26:2337–2346. doi: 10.1128/MCB.26.6.2337-2346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang KT, Wang J, Li LF, Su XD. Crystal structures of catalytic intermediates of human selenophosphate synthetase 1. J Mol Biol. 2009;390:747–759. doi: 10.1016/j.jmb.2009.05.032. [DOI] [PubMed] [Google Scholar]
- 26.Alsina B, Corominas M, Berry MJ, Baguñà J, Serras F. Disruption of selenoprotein biosynthesis affects cell proliferation in the imaginal discs and brain of Drosophila melanogaster. J Cell Sci. 1999;112:2875–2884. doi: 10.1242/jcs.112.17.2875. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.