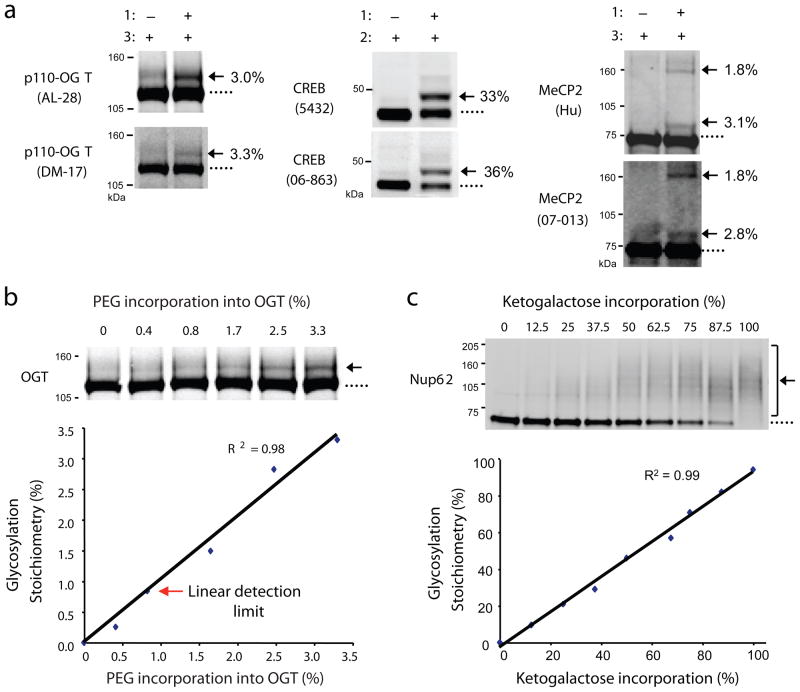
Figure 2.
Validation of the approach. (a) Distinct antibodies detect the same glycosylation stoichiometry on OGT (AL-28 and DM-17), CREB (Chemicon 5432 and Upstate 06-863), and MeCP2 (Hu and Upstate 07-013). p110-OGT was expressed in Sf9 cells, endogenous CREB was from rat liver, and MeCP2 was co-expressed with OGT in Sf9 cells. (b) As little as 0.8% of glycosylated OGT is readily detected. Sf9 cell lysate containing over-expressed p110-OGT was chemoezymatically labeled with PEG derivative 3 and diluted with unlabeled lysate to generate standards with varying percentages of label incorporation. The lysate was resolved by SDS-PAGE and immunoblotted with the anti-OGT antibody DM-17. The limit of detection was defined as the lowest stoichiometry value within 10% of the linear fit. See Supplementary Methods for details. (c) Detection of PEG incorporation into ketogalactose-labeled Nup62 is linear across a wide range of stoichiometries (0–100%). 293T cell lysate was labeled with UDP-ketogalactose 1 and then diluted with varying amounts of unlabeled lysate to simulate different levels of glycosylation. Each mixture was reacted with 2, resolved by SDS-PAGE, and immunoblotted for Nup62. Full-length blots are presented in Supplementary Figure 9.