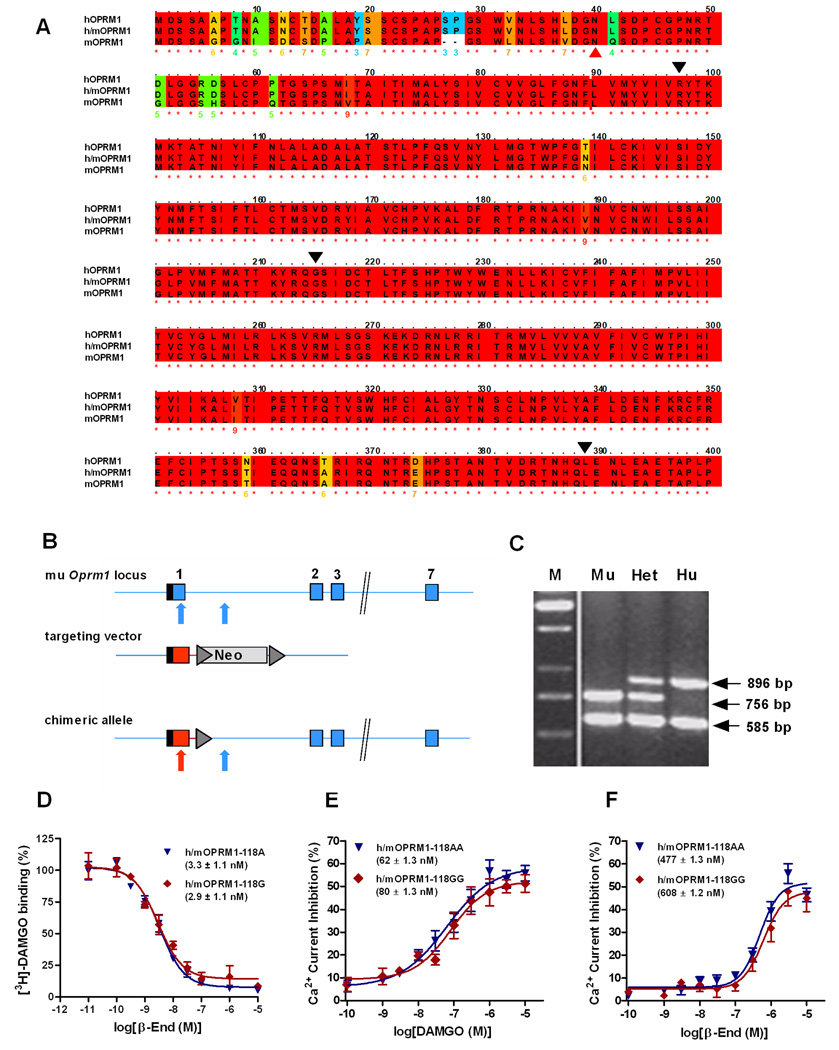
Fig. 2.
Targeting strategy and functional confirmation for humanized OPRM1 mouse lines. (A) Alignment of human (hOPRM1), mouse (mOPRM1) and humanized (h/mOPRM1) protein sequences. Black triangles indicate exon boundaries. Red triangle points to site of the N40D substitution, corresponding to A118G allelic variation. Color-coding of background indicates degree of conservation, with red indicating full human – mouse homology, and blue non-conservative substitution. As can be seen, replacement of mouse exon 1 with the corresponding human sequence yields a receptor that is highly homologous to the human protein. (B) Exon 1 of the murine OPRM1 locus including additional 400 bp of intron 1 was replaced by the corresponding human sequence, containing either A or G in position 118. Blue and red colors represent mouse and human sequences, respectively. Boxes: respective exons of the OPRM1 coding region. Hatched region: 5’-UTR. Triangle: frt site for flox recombinase. Neo: neomycin selection marker. Arrows point to position of the human and mouse specific PCR primers. (C) PCR based identification of the human (Hu), mouse (Mu) and heterozygote (Het) genotypes. M: Marker lane. (D) [3H]-DAMGO binding to respective humanized receptor transiently expressed in CHO cells. Data are means (±SEM) of triplicates, and mean (±SEM) IC50 values for the respective genotypes are indicated. Non-linear curve-fitting established that no significant genotypes differences were present (E – F) Concentration-response curves for DAMGO and β-endorphin (β-End) in trigeminal ganglion (TG) neurons isolated from h/mOPRM1 118AA or 118GG mice. Data points represent the mean (±SEM) of the agonist-mediated Ca2+ current inhibition. Mean EC50 values (±SEM) for the respective genotypes are indicated. Similar to the binding experiment, no genotype differences were found.