Abstract
Ethanol exerts complex effects on human physiology and health. Ethanol is not only addictive, but it is also a fetal teratogen, an adult neurotoxin, and an etiologic agent in hepatic and cardiovascular disease, inflammation, bone loss and fracture susceptibility. A large number of genes and signaling mechanisms have been implicated in ethanol's deleterious effects, leading to the suggestion that ethanol is a “dirty drug”. An important question is, are there cellular “master-switches” that can explain these pleiotropic effects of ethanol? MicroRNAs (miRNAs) have been recently identified as master regulators of the cellular transcriptome and proteome. miRNAs play an increasingly appreciated and crucial role in shaping the differentiation and function of tissues and organs in both health and disease. This critical review discusses new evidence showing that ethanol-sensitive miRNAs are indeed regulatory master-switches. More specifically, miRNAs control the development of tolerance, a crucial component of ethanol addiction. Other drugs of abuse also target some ethanol-sensitive miRNAs suggesting that common biochemical mechanisms underlie addiction. This review also discusses evidence that miRNAs mediate several ethanol pathologies, including disruption of neural stem cell proliferation and differentiation in the exposed fetus, gut leakiness that contributes to endotoxemia and alcoholic liver disease, and possibly also hepatocellular carcinomas and other gastrointestinal cancers. Finally, this review provides a perspective on emerging investigations into potential roles of miRNAs as mediators of ethanol's effects on inflammation and fracture healing, as well as the potential for miRNAs as diagnostic biomarkers and as targets for therapeutic interventions for alcohol related disorders.
Keywords: Tolerance, Fetal Alcohol Syndrome, Fetal Alcohol Spectrum Disorders, Neural Stem Cells, Alcoholic liver disease, hepatocellular carcinoma, Gastrointestinal Cancer, inflammation, bone fracture, miRNA, miR9, miR21, miR153, miR335, miR212, ZO-1
i. Overview
In the past decade or so, small non protein-coding RNAs (ncRNAs) have come to be appreciated as key regulators of gene expression. Among these, microRNAs (miRNAs) have garnered perhaps the most attention, as they are conserved among phylogenetically distant animals and are derived from previously unrecognized endogenous genes as well as from intronic and exonic regions of known protein-coding genes. Moreover, miRNAs have been shown to play critical roles in a broad array of fundamental biological processes, including regulation of the cell cycle, oncogenic transformation, stem cell regeneration and differentiation, immune cell differentiation, metazoan development, and organogenesis.
Consistent with these diverse roles, the regulatory potential of miRNAs is substantial as they number more than 700 in the humans alone (http://microrna.sanger.ac.uk; (Griffiths-Jones et al., 2008)) and often act as “master” regulators, capable of silencing the expression of large collections of target genes. These target genes are defined by short sequences in their 3' un-translated regions (UTRs) that are complementary to a given miRNA. Presumably, relegating these target sequences to this non-coding region has facilitated the expansion of the regulatory influence of miRNAs through evolution.
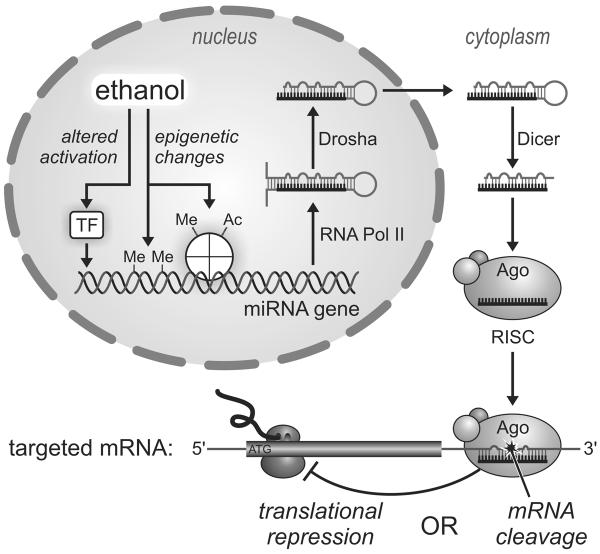
As depicted in Figure 1, primary miRNAs transcripts possess partial internal complementarity and, thus, adopt stem-loop hairpin structures. These precursors are then successively cleaved by two ribonucleases, Drosha and Dicer, to yield an RNA duplex, one strand of which is the mature 21–23-bp miRNA. In the cytoplasm, the miRNA associates with the Argonaute protein and additional accessory proteins to form a so-called RNA-induced Silencing Complex (RISC). Serving as an adaptor, the miRNA guides this effector complex to mRNAs possessing a complementary 3' UTR target sequence. Depending on the extent of complementarity, silencing typically results from either translational repression or Argonaute-catalyzed mRNA cleavage (Ambros, 2003).
Figure 1.
MicroRNA biogenesis and potential influences of alcohol. Hairpin primary miRNA transcripts are successively cleaved by RNases Drosha and Dicer while undergoing transport from the nucleus to the cytoplasm. Association of the mature miRNA with an Argonaut protein (Ago) directs the complex to complementary target sequences in specific messenger RNAs, leading to their cleavage or disruption of their translation. Ethanol may affect miRNA expression by altering activation of transcription factors (TF) and/or epigenetic modifications of DNA and DNA-associated histone complexes, including methylation (Me) and acetylation (Ac). See text for additional details.
i.a. Effect of ethanol on miRNA Expression and Function
Given the breadth of processes that miRNAs are known to regulate, it is reasonable to expect that they will play significant roles in mediating the effects of ethanol. While this remains largely unexplored, in the sections to follow, the authors present recent data showing that alterations in miRNA levels and miRNA-regulated biology are indeed associated with mechanisms of ethanol-induced tolerance, gut leakiness, and neural stem cell proliferation and differentiation (Pietrzykowski et al., 2008; Sathyan et al., 2007; Tang et al., 2008). In two additional recent reports (Dolganiuc et al., 2009; Wang et al., 2009a), using larger-scale miRNA screens, the authors reported that ethanol altered the expression of 2–3% of sampled miRNAs in murine models of ethanol-induced steatohepatitis and fetal brain injury. The miRNA changes observed were roughly equally divided between being up-regulated and down-regulated. While the absolute number of ethanol-sensitive miRNAs is apparently small, the capacity of individual miRNAs to control large numbers of genes suggests that miRNA expression changes will be amplified at the cellular and system level. Interestingly, only one miRNA, miR200a, was identified in both of these studies, suggesting that ethanol engenders tissue-specific miRNA responses, and hence tissue-specific miRNA-regulated biology.
i.b. Possible Mechanisms of Ethanol-induced Dysregulation of miRNA Expression
At this point we can only speculate about how ethanol exposure might trigger alterations in miRNA levels. It is well documented that ethanol alters the activities of various signal transduction pathways, including those involving receptor tyrosine kinases and MAP kinases, that regulate the activities of transcription factors and, thereby alter gene expression. Thus, it is entirely possible that ethanol can alter miRNA levels in this manner as well.
Another intriguing possibility worthy of consideration is that epigenetic changes due to ethanol may account for altered expression of some miRNAs. Epigenetic regulation of gene activity is primarily mediated by direct methylation of DNA itself as well as by acetylation, methylation, and phosphorylation of histone proteins, which govern DNA condensation. Several reports suggest mechanisms by which ethanol can reduce DNA methylation levels. Gestational ethanol exposure has been shown to cause reductions in fetal DNA methylation levels that could be explained by the ability of acetaldehyde, a metabolite of ethanol, to inhibit DNA methyltransferase activity (Garro et al., 1991). Ethanol consumption has also been shown to be associated with reduced DNA methyltransferase transcript levels and altered methylation of imprinted DNA regions in sperm (Bielawski et al., 2002; Ouko et al., 2009). In addition, chronic ethanol consumption can impair 1-carbon metabolism, consequently diminishing the availability of S-adenosyl-methionine (SAMe). This methyl donor is required for both DNA and histone methylation (Hamid et al., 2009; Shukla et al., 2008). Ethanol-mediated reductions in DNA methylation could be expected to increase expression of affected genes, including those encoding miRNAs.
The consequences of impaired histone methylation are less predictable as methylation may have opposing effects on gene regulation, depending which lysine residue is modified. However, a study of hepatocytes (Choudhury and Shukla, 2008) indicates that ethanol promotes histone H3 acetylation on lysine 9, a modification associated with transcriptional activation. This apparently results from a combination of histone acetyl transferase (HAT) activation and histone deacetylase (HDAC) inhibition. Taken together, these observations suggest that ethanol can alter the epigenetic landscape and thus contribute to altered regulation of miRNA expression.
II. miRNA roles in Ethanol Tolerance and Addiction
ii.a. miRNA contribution to ethanol tolerance
Drug tolerance is one of the initial steps leading to addiction, and an essential criterion in the diagnosis of drug dependence. Tolerance was initially described as a behavioral phenomenon, and defined as an increase in the exposure to a drug necessary to achieve constant response to the drug (Kalant, 1998). The American Medical Association identifies ethanol dependence (referred here also as alcoholism) as a psychiatric disease with characteristic signs and symptoms, and a progressive course (Hasin, 2003). About 12% of American adults will experience ethanol dependence at some time in their lives, incurring enormous social and medical costs. Tolerance to ethanol is an important inclusion criterion to diagnose alcohol dependence. The development of tolerance enforces increased ethanol consumption over time, to achieve the same level of intoxication. The principle of ethanol tolerance can also be observed at the molecular level. For example, initial exposure to ethanol activates an ion channel, however subsequent chronic ethanol exposure may abolish activation of that channel. Modern life sciences assume that molecular mechanisms underpin every type of behavior. However, the exact mechanisms underlying molecular tolerance are complex, and far from well understood (Pietrzykowski and Treistman, 2008).
We recently discovered that a miRNA-based mechanism contributes to the development of molecular tolerance to ethanol (Pietrzykowski et al., 2008). We observed that in rodent neurons, isolated from the central nervous system, ethanol upregulates one particular miRNA, miR9, which subsequently affects expression of mRNA splice variants of the main, pore-forming, alpha subunit of BK channel, a high conductance calcium- and voltage-dependent potassium channel (Atkinson et al., 1991). We determined that neurons co-express multiple mRNA variants encoding different isoforms of the BK channel, which vary in some of their properties. Expression of several isoforms of the same ion channel presumably enables a cell to be “plastic” and to respond quickly to everchanging environmental cues. Interestingly, BK channel isoforms also differed in their activation by ethanol. Some isoforms were very sensitive to ethanol, while others have low sensitivity or even innate tolerance to ethanol. We hypothesized that the molecular mechanism of development of ethanol tolerance by the BK channel will include a change in the ratio of ethanol sensitive and ethanol tolerant isoforms towards ethanol tolerant isoforms. Quantification of individual variants before ethanol exposure and at various times of exposure (ranging from 15 min to 24 hrs) to physiologically relevant ethanol concentrations (20 mM) indeed supported our hypothesis. We observed a radical reduction of splice variant diversity - specifically a profound decrease in ethanol-sensitive variants. Computational modeling indicated that this decrease contributed to a relative increase in the expression of mRNA variants encoding ethanol tolerant BK channels.
Recent discoveries point to miRNAs and other ncRNAs as fundamental regulatory molecules. Therefore, we decided to determine whether miRNAs could play a role in the described mechanism of the development of molecular tolerance to ethanol. To answer this question we focused on the BK channel 3'UTR – an untranslated region of mRNA, known for its regulation of mRNA stability and a target for miRNAs. We discovered that the BK channel possess several alternate 3'UTRs, each exhibiting different miRNA binding patterns. Interestingly, a miR9 binding site was present on only one of these 3'UTRs. Further experiments indicated that the 3'UTR containing a miR9 binding site is “stitched” to mRNA transcripts encoding BK isoforms of high ethanol sensitivity. Thus, ethanol by upregulating miR9, increases the probability of interaction between miR9 and its binding site located on specific 3'UTRs. As a consequence of this interaction, mRNAs associated with these 3'UTRs are degraded, biasing the ratio of ethanol sensitive/ethanol tolerant variants toward tolerance.
The BK channel is abundant in brain and is critical for neuronal function (Dworetzky et al., 1994). However, there is mounting evidence that miRNAs influence multiple targets. Therefore, in the last series of experiments, we searched whether a similar regulation can be attributable to other molecules important for neuronal activity and known to be affected by ethanol exposure. Indeed, our data indicate that ethanol via miR9 can also affect expression of at least ten other molecules crucial for synaptic plasticity, circadian rhythm, or neurotransmitter release (Pietrzykowski et al., 2008).
ii.b. Alcoholism and miRNAs
Alcoholism is a multigenic disease, in which large gene networks are affected. Considering widespread effect of miR9 on expression of several genes relevant to ethanol actions on the CNS, it is tempting to speculate that miR9 plays a substantial role in mediating ethanol's effects on the development of alcoholism. The mammalian genome contains three independent copies of the miR9 gene (miR9-1, 9-2 and 9-3 on human chromosomes 1, 5 and 15 respectively, according to miRbase, (Griffiths-Jones et al., 2008)). Although the mature form of each miR9 gene-derived miRNA is identical, there are substantial differences in their precursors as well as promoter regions. It is worth investigating whether this genomic variability contributes to the expression of miR9, and whether there are specific changes in miR9 genes and their regulatory regions in humans addicted to ethanol.
ii.c. miRNAs and other drugs of abuse
“miRNA-ology” is a relatively young field and publications describing roles of miRNAs in addiction are starting to emerge. Besides ethanol, a role for miRNAs has recently been assessed for two other drugs of abuse, nicotine and morphine.
Recently, Huang and Li (Huang and Li, 2009) tested the hypothesis that miRNAs mediate effects of nicotine on gene expression. Using rodent neuronal cell culture and miRNA microarrays they measured the effect of 1 hour of 100 uM nicotine exposure on miRNA expression. They determined that short-term nicotine exposure upregulated the expression of 11 miRNAs, while downregulating the expression of an additional 14 miRNAs. Next, they focused on miR140* because it potentially targeted several genes important for neuronal function. One of the prominent targets of miR140* is Dnm1. This gene encodes a large GTPase, dynamin-1, which plays a central role in synaptic endocytosis (Ferguson et al., 2007). Huang and Li showed in a series of experiments that expression of miR140* is upregulated by nicotine exposure, that miR140* can bind to Dnm1mRNA 3'UTR region, and that miR140* regulates expression of Dnm1 mRNA. Specifically relevant to research on ethanol addiction, these authors also report that nicotine downregulates miR21 and miR335. Interestingly, as discussed below (Sathyan et al., 2007), ethanol also down-regulates these two miRNAs. Since ethanol is frequently co-abused with nicotine, these data point toward an intriguing possibility that at least some of miRNAs can explain overlapping mechanisms of ethanol and nicotine actions.
Chronic nicotine (Shan et al., 2009) has also recently been shown to decrease the expression of miR133 and miR590 in canine cardiomyocytes, while targets of these two miRNAs, TGF-β1 and TGF-βRII, were upregulated. As TGFs play an important role in regulating the production of collagens and the development of myocardial fibrosis, nicotine regulation of miRNAs could contribute to nicotine-related atrial remodeling and arrhythmias. Ethanol is well known to cause severe heart remodeling and malfunctions e.g. cardiomyopathy, supraventricular arrhythmias (Klatsky, 2009). It is tempting to hypothesize that, similarly to nicotine, ethanol-related heart failure is caused, at least partially, by miRNA-dependent mechanisms.
Evidence that miRNAs are also involved in actions of opioids in neurons comes from work by Wu (Wu et al., 2008b; Wu et al., 2009). They discovered that the 3'-UTR of the mu-opioid receptor (MOR1) mRNA contains a functional binding site for the miRNA miR23b. miR23b binds to MOR1 3'UTR via a K box motif halting association of the mRNA with polysomes and arresting MOR1 protein production, without causing mRNA degradation (Wu et al., 2008b). Moreover, prolonged exposure to morphine caused a dose-dependent increase in miR23b levels in neuronal cells, suggesting a role of miR23b in an auto-regulatory feedback loop. Interestingly, miR23b upregulation was only observed after exposures longer then 6 h (Wu et al., 2009). Further studies are warranted to determine the exact molecular mechanisms underlying this phenomenon.
ii.d. miRNAs and drug addiction
The role of miRNAs in drug addiction is just starting to be investigated. Pioneering work in this field indicates clearly that miRNAs play important roles in the actions of ethanol and other drugs of abuse. The emerging picture is already very complex, showing an intricate relationship between miRNAs and drugs of abuse. Ethanol can cause simultaneous upregulation of some miRNAs while downregulating others. Ethanol's effect on a particular miRNAs can depend on dose and cell type. Some miRNAs can be modulated by both ethanol and nicotine, suggesting a novel overlap in the molecular mechanisms of these drugs. However, a clear outcome of these initial studies is the evidence that drugs of abuse can control the expression of genes and gene networks in cells, by controlling miRNAs. Perhaps miRNAs serve as molecular convergence points for mechanisms that underlie the development of drug tolerance and dependence.
III. Ethanol's Effects on Neural Stem Cell Maturation: Implications of miRNAs for Fetal Ethanol Teratology
Ethanol is clearly addictive and as discussed above, miRNAs can control important components of addiction like the development of tolerance (Pietrzykowski et al., 2008). However, ethanol is also an important teratogen, and emerging evidence shows that miRNA expression is sensitive to ethanol during development and that miRNAs mediate ethanol teratology (Sathyan et al., 2007; Wang et al., 2009a).
iii.a. Ethanol as a teratogen
Heavy ethanol consumption during pregnancy can lead to growth retardation, mental retardation, and a constellation of craniofacial, cardiovascular and skeletal defects that are collectively termed the “Fetal Alcohol Syndrome” or FAS (Clarren, 1986; Clarren et al., 1978). The rate of FAS births in the general population has not declined (estimated at ~0.2–1.5 cases/1000 live births (Bertrand et al., 2005)), and the rates of Fetal Alcohol Spectrum Disorders (FASD, a broader category that includes less severe ethanol effects, (Sokol et al., 2003)) are unknown, but estimated to be significantly higher. Ethanol consumption during pregnancy continues to be the leading preventable cause of childhood mental retardation in the United States.
iii.b. Fetal stem cells are an ethanol target
The tail-end of the first trimester, and the second trimester of pregnancy together represent a unique period of fetal vulnerability to ethanol and other teratogens, because, during this period of development, which corresponds to the second half of gestation in the mouse and rat, stem cells (NSCs) and progenitor cells (NPCs) of the fetal ventricular and subventricular zones proliferate rapidly to generate most of the neurons of the adult brain (Bayer et al., 1993). Second trimester-equivalent exposure to ethanol has been reported to lead to a loss of hippocampal neurons (Barnes and Walker, 1981) altered cortical neurogenesis (Miller, 1989; Miller, 1996; Miller and Nowakowski, 1991), enlargement of the sub ventricular zone, an intermediate neurogenic layer (Kotkoskie and Norton, 1989), disorganized cortical architecture (Kotkoskie and Norton, 1988), and a disruption in lamination patterns specifically in the earliest generated lamina of the emerging cortical plate (Kotkoskie and Norton, 1989).
In a series of experiments using fetal rodent neurosphere cultures to model the second trimester fetal neuroepithelium, we found that fetal NSCs/NPCs were a direct target of ethanol. Surprisingly both rodent (Prock and Miranda, 2007; Santillano et al., 2005) and human (Vangipuram et al., 2008) fetal derived NSCs/NPCs are highly resistant to ethanol-induced cell death, even at doses up to 200 mM. However, we observed that ethanol, at abuse-relevant doses, induced an increase in the size of neurosphere cultures, and increased in the proportion of cells in S and G2/M phase (Santillano et al., 2005). This ethanol-induced increase in cell cycle was accompanied by the loss of a specific stem cell-specific marker (ABCG2) as well as a loss of other stem/progenitor markers (CD117, CD133 and Sca-1), without an overall change in the expression of the generic immaturity marker, nestin. From these data, we hypothesized that ethanol, promoted cell cycle, resulting in increased maturation, and consequently, depletion of stem and early progenitor cells. In support of this hypothesis, we observed that ethanol-pretreated neurosphere cultures exhibited increase evidence for morphologic asymmetric cell division, and an emergence of radial glial-like cells. Since radial glia are the immediate precursors for neurons (Anthony et al., 2004; Gotz and Barde, 2005), these data lend further support to a hypothesis that ethanol depletes neural stem cell pools by inducing maturation rather than death. A final piece of evidence that ethanol is teratogenic to stem cells, comes from our observation that differentiating neuroblasts, derived from ethanol pre-exposed neurosphere cultures exhibit significantly increased migration, compared to non-exposed controls, when cultured on a permissive extracellular matrix substrate (Camarillo and Miranda, 2008). This final evidence for ethanol teratogenesis in NSCs/NPCs is important, as this is direct evidence for persistent (organizational) effects of ethanol in NSCs/NPCs. Additionally these data also advance a possible mechanism for why ethanol exposure can lead to the formation of fetal neuronal heterotopias in both children with FAS (Clarren, 1986) and in second trimester rodent models (Komatsu, 2001; Mooney et al., 2004).
iii.c. MiRNA targets of ethanol control fetal neural stem cell maturation
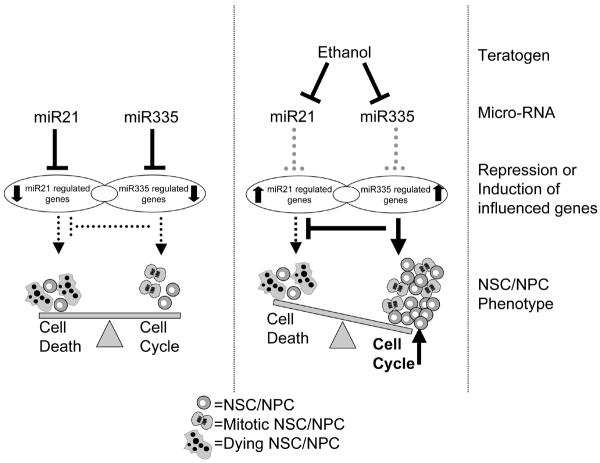
The effects of ethanol on NSC/NPC maturation are ultimately complex and, as the above data indicate, likely to result from the mis-recruitment of large gene networks and complex biological processes. This factor reinforces the notion that ethanol is a “dirty drug” with pleiotropic effects on cells and tissues, and stymies efforts to develop a coherent model for ethanol teratogenesis. Nevertheless, the identification of miRNAs as biological targets of ethanol offers one hope for developing a more coherent model of ethanol's actions, because individual miRNAs have the capacity to control large gene networks to promote complex biological endpoints. In a preliminary screen, we identified four miRNAs, miR9, miR21, miR153 and miR335, that were suppressed by ethanol in NSCs/NPCs (albeit at a relatively high dose, 70mM, more likely to be attained in an alcoholic (Adachi et al., 1991)). We observed that knocking down miR21 resulted in rapid induction of apoptosis. However, concurrently knocking down miR335 prevented the cell death resulting from knockdown of miR21. These data indicated that miR21 was anti-apoptotic, whereas miR335 in contrast, was a pro-apoptotic miRNA. Recent evidence from other laboratories shows that miR153 is also a pro-apoptotic miRNA, because it suppresses the expression of anti-apoptosis genes like Bcl-2 (Xu et al., 2009). Therefore, ethanol's concurrent suppression of miR21, miR153 and miR335 explains the apoptosis resistance of ethanol-exposed NSCs/NPCs. MiR335 knockdown also specifically resulted in increased BrdU incorporation in neurosphere cultures, indicating that miR335 suppression may be the mechanism for ethanol-induction of cell proliferation in neurosphere cultures (Figure 2).
Figure 2.
Model for the collaborative role for miR21 and miR335 in shaping fetal NSC/NPC fate in the presence and absence of ethanol. Mir21 is anti-apoptotic, while miR335, its functional antagonist, is pro-apoptotic (see text for details). The model shows that in the absence of ethanol, miR21 and miR335 cooperatively repress gene expression to balance cell survival and proliferation. Ethanol decreases both miRNAs (along with miR153 and miR9), resulting in derepression of gene expression, and consequently, increased cell proliferation while maintaining overall apoptosis resistance. We hypothesize that this increase in proliferation following miR-335 suppression prematurely depletes stem cells from the fetal neuroepithelium.
In silico analyses predicted that the Notch receptor ligand, Jagged-1, and the neuron-specific RNA binding protein ELAVL2/HuB were each targets of three out of four ethanol-sensitive miRNAs respectively. As predicted, mRNA transcripts for both Jagged-1 and ELAVL2/HuB were both induced by ethanol in neurosphere cultures. ELAVL2/HuB over-expression is sufficient to promote neuronal differentiation (Akamatsu et al., 1999). The role of Jagged-1 is more complex. Though activation of the notch pathway is associated with NSC proliferation (Basak and Taylor, 2007), Jagged-1-induced proliferation helps establish neuronal identity (Salero and Hatten, 2007). The induction of predicted differentiation-associated genes would therefore, seem to further support a hypothesis that ethanol promotes NSC maturation by de-repressing miRNA-inhibited neuronal identity factors.
iii.d. Developmental and evolutionary constraints on miRNA involvement in the teratology of ethanol
Interestingly, while ethanol suppresses miR9 in fetal-derived NSCs/NPCs (Sathyan et al., 2007), miR9 is induced by ethanol in adult neurons (Pietrzykowski et al., 2008). Clearly, developmental stage is likely to be a critical modulator of ethanol effects in target tissues. An important and unanswered question is how developmental stage alters ethanol's actions. Mir9 is an example of the potential complexity that besets the study of miRNA targets of ethanol. The mammalian genome contains three independent copies of the miR9 gene (as mentioned above). While the mature copies of miR9 are identical to each other, the premature miRNA transcripts and their gene loci exhibit substantial variability. Emerging evidence suggests that miR9 plays a role in maintaining neural stem cell fate by modulating transcription and epigenetic factors (Li et al., 2006; Shibata et al., 2008; Zhao et al., 2009). Further investigations will be required, to determine if different miR9 genes exhibit distinct spatial and temporal expression patterns, and whether development stage-specific transcription and epigenetic environments in turn influence the promoters for different miR9 genes.
While miR9 is expressed throughout vertebrate evolution, miR335 first appeared in eutherian, (i.e., true placental) mammals, as an intronic miRNA within the maternally imprinted gene MEST/Peg1 (UCSC genome browser, http://genome.ucsc.edu/, (Kent et al., 2002)). While we know little about miR335's function, maternal uniparental disomy (duplication of the imprinted maternal chromosome and loss of the expressed paternal chromosome locus) in the region encompassing the MEST/Peg1, is an etiologic factor in the Russell-Silver syndrome, an intrauterine and postnatal growth retardation syndrome, characterized by developmental cognitive delay (Kotzot, 2008), with features somewhat similar to that observed in FAS. Moreover, inactivation of the paternal MEST/Peg1 locus leads to fetal and postnatal growth retardation in mouse models (Lefebvre et al., 1998). Finally, the MEST/Peg1 locus is also implicated in FGF-dependent rostro-caudal patterning of the fetal cerebral cortex (Sansom et al., 2005). While the specific contribution of miR335 to fetal growth and to fetal cortical patterning is currently unknown, miR335 (along with miR9) has recently been shown to be suppressed during the maturation of A2B5-postitive oligodendrocytic progenitors (Lau et al., 2008), suggesting that it may have a role, together with miR9, in preventing the maturation of neural progenitors. It is tempting to speculate that miR335 plays a mammalian-specific role in brain development given its evolutionarily recent, mammalian-specific expression within fetal NSCs/NPCs, its capacity to control cell survival and proliferation, and its potential to repress differentiation. Consequently, miR335's sensitivity to ethanol suggests that this miRNA in conjunction with more evolutionarily conserved miRNAs like miR9, miR21 and miR153 and perhaps others more recently discovered (Wang et al., 2009a), may confer mammalian-specific patterns of sensitivity to teratogens like ethanol.
IV. MiRNAs in ethanol-induced gastrointestinal disease
iv.a. A nexus between alcoholic liver disease (ALD) and ethanol-induced gut epithelial dysfunction
Mounting evidence suggests that aberrant miRNA expression is associated with several ethanol-induced gastrointestinal (GI) diseases, including ALD, a common and serious complication of heavy drinking (Burbige et al., 1984; Galambos, 1972; Grant et al., 1988; Maher, 2002). Since only ~30% of alcoholics develop ALD (Grant et al., 1988), additional factors besides excessive ethanol consumption must be involved. Indeed, several recent clinical observations and experimental studies strongly suggest that gut-derived endotoxin is the key cofactor for ALD (Adachi et al., 1995; Bhagwandeen et al., 1987; Bode et al., 1987; Keshavarzian et al., 1999; Nanji et al., 1994). Our previous studies showed that only alcoholics with ALD, and not those without ALD, developed intestinal hyper-permeability (Keshavarzian et al., 1999). We also showed that chronic daily ethanol administration to rats caused gut leakiness through iNOS signaling (Keshavarzian et al., 2001; Keshavarzian et al., 2009; Tang et al., 2009b). Importantly, ethanol-induced gut leakiness in rats was associated with endotoxemia and alcoholic steatohepatitis (Keshavarzian et al., 2009), and oats supplementation prevented loss of intestinal barrier integrity, endotoxemia and steatohepatitis (Keshavarzian et al., 2001; Tang et al., 2009a). These findings suggesting that gut leakiness is permissive for the endotoxemia associated with ALD are not surprising. The intestinal epithelium is a highly selective barrier that permits the absorption of nutrients from the gut lumen into the circulation, but, normally, restricts the passage of harmful and potentially toxic compounds including products of luminal microbiota (Clayburgh et al., 2004; Hollander, 1992; Keshavarzian et al., 1999). Disruption of intestinal barrier integrity (leaky gut) may lead to the penetration of bacterial products such as endotoxin from the lumen into the mucosa, and then into the systemic circulation, and initiate local inflammatory processes in the intestine and even in distant organs such as the liver (Clayburgh et al., 2004; Hollander, 1992; Keshavarzian et al., 1999).
The primary epithelial structures regulating the intestinal barrier are tight junctional [TJ] and adherens junctional proteins. Tight junctions function as gates that regulate intestinal permeability (Clayburgh et al., 2004; Ivanov et al., 2005; Laukoetter et al., 2006; Turner, 2006). One key TJ protein is zonula occludens 1 (ZO-1), and ethanol-induced dysregulation of ZO-1 in intestinal epithelial cells could underlie ethanol-induced gut leakiness. The question is how chronic ethanol consumption disrupts TJ proteins including ZO-1.
iv.b. miRNA involvement in ethanol-mediated gut epithelial dysfunction
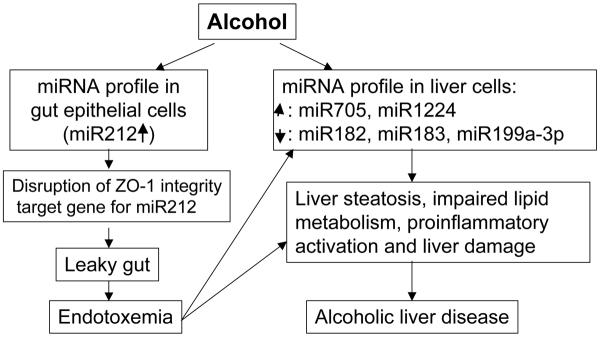
We recently explored the possibility that miRNAs are involved in the disruption of ZO-1, hypothesizing that ethanol-induced gut leakiness could be due to the effects of ethanol on expression of miRNAs that target TJ genes in intestinal epithelial cells. We studied the effects of ethanol on miR212 in intestinal epithelial cells because preliminary in silico analyses (Blow et al., 2006; Griffiths-Jones et al., 2006) predicted that ZO-1 was a target of miR212. We first showed (Tang et al., 2008) that miR212 is highly expressed in intestinal tissues. We also showed that ethanol-induced miR212 over-expression is accompanied by reductions in ZO-1 protein expression, disruption of tight junction protein (ZO-1), and increased permeability of monolayers of Caco-2 cells. Ethanol-induced miR-212 over-expression correlated with ethanol-induced disruption of monolayer integrity. Most importantly, we found (Tang et al., 2008) that miR212 levels in colon biopsy samples in ALD patients were higher than in healthy controls; ZO-1 protein levels were lower. To see if miR212 regulates ZO-1 levels, we conducted both overexpression studies using miR212 precursors and inhibition studies using miR212-specific antisense oligonucleotide inhibitors (anti-miR212). The data showed that miR212 over-expression significantly inhibited ZO-1 protein expression and knocking down of miR212 expression in Caco-2 cells using anti-miR212 significantly inhibited ethanol-induced hyperpermeability by 50% (Tang et al., 2008). These studies suggest a novel mechanism for ethanol-induced gut leakiness, namely, that ethanol induces miR212 over-expression, which disrupts intestinal barrier integrity by inhibiting ZO-1 translation. This cascade leads to dysfunction of tight junctions and increased intestinal permeability (Figure 3). This mechanism provides a potential therapeutic target for preventing gut leakiness in patients with ALD (Tang et al., 2008).
Figure 3.
Model for miRNA involvement in Alcoholic Liver Disease (ALD). ALD can result from both direct actions of ethanol on the liver and indirectly, from ethanol's actions on the enteric epithelium. The indirect mode of action implicates miRNA dysregulation in the gut as a permissive agent for ALD. For example, the ethanol-mediated increase in miR212 leads to increased gut leakiness by disrupting the expression of key enteric tight junction scaffolding proteins like ZO-1. The resulting endotoxemia initiates hepatic damage and may lead indirectly to the development of ALD. The composite model is based on research by Tang et al., 2008, and Dolganiuc et al., 2009. See text for additional details.
Ethanol-induced changes in miRNA profiles in the digestive system are not limited to gut leakiness. For example, ethanol-induced changes in miRNA expression in the liver was recently reported (Dolganiuc et al., 2009). These authors found an aberrant liver miRNA profile in alcoholic steatohepatitis (ASH induced by feeding mice a Lieber-DeCarli ethanol diet) and non-alcoholic steatohepatitis (NASH induced by feeding mice a methionine-deficient (MCD) diet). They reported that the Lieber-DeCarli diet up-regulates ~1% and down-regulates ~1% of known miRNAs; the MCD diet up-regulates ~3% and down-regulates ~1% of known miRNAs in mouse liver. After 5 weeks of either diet, expression of both miR705 and miR1224 was increased, while expression of miR182, miR183, and miR199a-3p was down-regulated during Lieber-DeCarli feeding and upregulated during MCD feeding. The authors proposed that changes in miRNAs contribute to impaired hepatic lipid homeostasis and the inflammatory cascade in ASH and NASH (Figure 3).
iv.c. A role for miRNAs in ethanol-induced GI cancer
Lineage differentiation of intestinal epithelial cells is determined by developmental and environmental signals. A role for miRNA in the differentiation of intestinal epithelial cells was recently reported (Hino et al., 2008). This study showed that miR194 is highly induced by hepatocyte nuclear factor-1 (HNF-1), a transcription factor that regulates gene expression in intestinal epithelial cells. Their studies indicated that miRNAs are involved in lineage commitment decisions in intestinal epithelium and that miRNA are regulated in a tissue-specific manner in intestinal epithelium (Hino et al., 2008). Not surprisingly, dysregulated miRNA profiles have been reported for several GI cancers such as colon and liver cancers. For example, the role of miRNAs in colon cancer was first reported by Michael et al (Michael et al., 2003). They identified miR-143 and miR-145 as dysregulated miRNAs in colon cancer. Since then, the role of miRNAs in the carcinogenesis, genomics, prognosis, and chemotherapy of colon cancer were studied and reviewed by others (Yang et al., 2009). Recently, Sung et al reported that differential expression of miRNAs in plasma of colorectal cancer patients is a potential marker for colorectal cancer screening (Ng et al., 2009a). Tryndyak et al (Tryndyak et al., 2009) studied the role of miRNAs in rat model for dietary methyl-deficiency induced hepatocarcinogenesis; this is relevant to hepatocarcinogenesis in humans that is associated with viral hepatitis B and C infection, and with alcoholic and metabolic liver diseases. They showed that methyl-deficiency inhibited the expression of miRNAs, miR34a, miR127, miR200b, and miR16a, which are involved in the development of hepatocellular carcinoma (HCC) through regulation of apoptosis, cell proliferation, intercellular adhesion, and epithelial-mesenchymal transitions (Tryndyak et al., 2009). miRNA expression profiling in human HCC cells and tissues and in experimental models of HCC have been reviewed (Braconi and Patel, 2008). Recently, Kota et al (Kota et al., 2009) reported that HCC cells exhibit reduced expression of miR26a, an miRNA that is normally expressed at high levels in diverse tissues. They induced miR26a expression through systemic administration of miR26a in a mouse model of HCC using adeno-associated virus. Their results showed that induction of miR26a expression leads to inhibition of cancer cell proliferation, induction of tumor-specific apoptosis, and dramatic protection from disease progression without toxicity. These findings suggest that therapeutic strategies based on modulation of miRNA activity hold great promise for many disorders in which miRNAs play an important role in the disease mechanism (Kota et al., 2009). Thus, It is highly plausible that ethanol-induced changes in miRNA profiles are involved in the increased incidence and in the clinical course of several GI cancers. Proof of this hypothesis requires further studies.
iv.d. miRNAs and inflammatory GI disease
Finally, changes in miRNAs that are induced by ethanol could be key in mediating ethanol-induced inflammatory processes in intestine and liver because the potential importance of miRNAs in intestinal inflammation has recently been demonstrated. Wu et al (Wu et al., 2008a) measured miRNA expression in the intestinal mucosa of patients with active ulcerative colitis (UC), inactive UC, Crohn's disease, irritable bowel syndrome, infectious colitis, and microscopic colitis, as well as in healthy subjects by microarray and RT-PCR. Their results showed that 11 miRNAs were differentially expressed in active UC (8 increased and 3 decreased significantly). miR192 expression was decreased in colonic epithelial cells in active UC. Macrophage inflammatory peptide (MIP)-2 was identified as a target of miR192. In colonic epithelial cells, TNF, a proinflammatory cytokine that is present in high levels in the inflamed intestine, induced miR192 reduction and MIP-2 overexpression (Wu et al., 2008a). MiRNAs not only appear to be involved in gut inflammation, but also, in regulation of intestinal functions such as fluid secretion. For example, Kapeller et al reported an association between a functional variant of miR510 target site of the serotonin receptor-type 3E gene (HTR3E) and diarrhea predominant irritable bowel syndrome (Kapeller et al., 2008). It remains to be seen whether miRNAs that target inflammation regulatory genes also play a role in ethanol-induced inflammation in the intestine and liver.
In summary, recent studies provide compelling evidence for the role of miRNAs in mechanisms underlying several ethanol-induced GI disorders. MiRNAs therefore represent new therapeutic targets for the prevention and/or treatment of ethanol-related intestinal and liver diseases.
v. Emerging Perspectives
v.a. MiRNAs and ethanol, implications for inflammation and fracture healing
Orthopedic surgeons have long recognized that fractures of alcoholics are more difficult to heal successfully (Nyquist et al., 1997; Tonnesen et al., 1991) and have a higher incidence of non-union3 (Foulk and Szabo, 1995; Nyquist et al., 1998), perhaps due to perturbations in the initial, inflammatory stage of fracture healing. The inflammatory stage of fracture healing involves a rapid and short lived influx of neutrophils, followed by monocyte/macrophage infiltration and finally lymphocytes. Each of these cell types migrate to the site and are maintained under the direction and signaling of chemokines and other regulatory factors. Inflammation involves a highly coordinated sequence of events controlled by positive and negative mechanisms, most assurdaly including miRNAs.
Chronic ethanol consumption dysregulates the immune system, favoring Th2- over Th1-type immune responses, impairing the function of neutrophils and monocytes (Szabo, 1999), cells that also control the inflammatory stage of fracture healing. An analysis of the involvement of miRNAs in Th1-type inflammatory responses may therefore inform us about the potential role of miRNAs in fracture healing as well. Bacterial endotoxin has recently been shown to induce several miRNAs, e.g., miR132, miR146a/b and miR155, in a human monocyte cell line (Taganov et al., 2006). MiR155 was also induced in bone marrow-derived macrophages, stimulated via a variety of different mechanisms and pathways, including ethanol-sensitive signaling molecules like TNF-α, JNK, and AKT (Androulidaki et al., 2009; O'Connell et al., 2007; Tili et al., 2007), suggesting that miR155 is a common target of the primary macrophage response to a broad range of inflammatory mediators. Interestingly, endotoxin-mediated activation of mouse bone marrow, results in an induction of miR155 as well as granulocyte/monocyte expansion (O'Connell et al., 2008), and miR155 is overexpressed in the bone marrow of patients with certain types of acute myeloid leukemia (O'Connell et al., 2008), suggesting that miR155, once induced, contributes to the expansion of myeloid lineages.
Other known ethanol sensitive miRNAs like miR9 may also contribute to inflammation-associated activation of myeloid cells. For example, a recent report (Bazzoni et al., 2009) showed that miR9 was up-regulated in human neutrophils and monocytes, when stimulated by TLR4 or the proinflammatory cytokines TNF-α, and IL-1β. A final interesting miRNA involved in blood cell production and the inflammatory response is miR223, which is expressed nearly exclusively in bone marrow (Chen et al., 2004). MiR223 increases as granulocytic differentiation proceeds from granulocyte-monocyte progenitors through peripheral blood granulocytes, but is repressed when the progenitors adopt the monocyte fate (Johnnidis et al., 2008). Mef2c, a transcription factor that promotes myeloid progenitor proliferation, is a target for miR-223 (Johnnidis et al., 2008). Overexpression of miR223 doubled the cells committed to the granulocyte-specific lineage while knock-down had the opposite effect (Fazi et al., 2005). They also demonstrated regulatory control of miR233 by the transcriptional factors NFI-A and C/EBPα, with NFI-A maintaining it at low levels and C/EBPα up-regulating its expression (Fazi et al., 2005). The involvement of miRNAs in the recruitment of neutrophils and monocytes to support inflammatory responses suggests a strong likelihood that these miRNAs will also play an important role in fracture healing.
v.b. MiRNA profiling in postmortem brain of human alcoholics
A number of studies have used gene expression profiling to identify differentially expressed genes in postmortem brain of long-term alcohol abusers (Flatscher-Bader et al., 2005; Iwamoto et al., 2004; Lewohl et al., 2000; Liu et al., 2004; Liu et al., 2006; Mayfield et al., 2002; Sokolov et al., 2003). The prefrontal cortex, which is important for judgment, decision-making, and other cognitive function, has been of particular interest in several expression studies because of its susceptibility to damage by ethanol abuse (Harper et al., 1985; Kril et al., 1997; Kril and Harper, 1989). In a previous study we investigated 27 individual cases (14 well characterized alcoholics and 13 matched controls) and found that long-term ethanol abuse altered the expression of a number of functionally related families of genes including myelination, ubiquitination, apoptosis, cell adhesion, neurogenesis, and neural disease (Liu et al., 2006). Importantly, gene expression patterns were identified that accurately discriminated between control and alcoholic cases suggesting that ethanol abuse reprograms gene transcription in specific and potentially predictable ways. It is interesting to speculate that the genes that are important in distinguishing control from alcoholic groups are under the control of regulatory elements such as miRNAs.
As discussed in previous sections, miRNAs can target many mRNA transcripts for either translation repression or degradation, but the function of many human miRNAs is unknown. Since altered gene expression patterns have been identified in response to long-term ethanol consumption it is possible that miRNAs play a role in this regulation. While few published miRNA reports have focused on human brain, support for this hypothesis comes from recent studies demonstrating that peripheral myelin protein 22 (PMP22) is a target of miR29a suggesting that myelin gene expression is regulated in part by miRNAs (Verrier et al., 2009). Long-term ethanol use has been shown to significantly regulate a number of myelination-related genes including PMP22 in a number of postmortem brain studies (Flatscher-Bader et al., 2005; Lewohl et al., 2000; Liu et al., 2006). In addition, there is growing evidence that altered miRNAs may be involved in a variety of neurodegenerative disorders including Alzheimer's, Parkinson's, and prion diseases (Barbato et al., 2009; Hebert and De Strooper, 2009; Hebert et al., 2009; Junn et al., 2009; Montag et al., 2009). Interestingly, families of genes involved in neurodegeneration are significantly overrepresented in the prefrontal cortex of alcoholics (Liu et al., 2006) suggesting a link between alcoholism and other neurodegenerative conditions. Together these studies raise the possibility that the altered expression of brain genes observed in human alcoholism may be due to ethanol-induced changes in the levels of miRNAs.
v.c. Diagnostic and therapeutic applications of miRNAs in ethanol abuse and dependence
Early diagnosis and treatment of complex diseases such as alcoholism may benefit from miRNA studies designed to identify disease-specific molecular markers that provide a fingerprint of the condition, or to identify potential therapeutic targets. Rapid advances in the field of genomics offer new diagnostic and screening potential even for complex genetic diseases like alcoholism. The importance of understanding miRNA expression changes in ethanol abuse and dependence can be appreciated by the impact of numerous studies in other diseases, most notably cancer, where studies have led to improved therapeutic strategies (Kota et al., 2009; Ng et al., 2009b; Rossi, 2009) and to a molecular classification of disease that promises to be more accurate and informative than traditional diagnostic tests (Heneghan et al., 2009; Wang et al., 2009b). A better understanding of ethanol-induced changes in miRNA expression may also provide an accurate means to diagnose various consequences of ethanol abuse as well as to identify potential therapeutic strategies to treat the condition and/or the consequences of heavy ethanol use.
VI. Summary
The studies outlined in this review highlight a number of important findings that suggest that miRNAs play key roles in regulating genes involved in a variety of biological processes, such as cell proliferation, inflammation, neuronal differentiation, developmental timing, synapse function, and neurogenesis. Given that a single miRNA can potentially target hundreds of mRNA transcripts for either translation repression or degradation, it is possible that these small RNAs serve as “master” regulators of cell function. In addition, these studies demonstrate that the regulatory potential of miRNAs is not restricted to specific cell types, model systems, or species underscoring the potential role of miRNAs in orchestrating the complexities involved in normal cellular function.
These studies also demonstrate that ethanol and other drugs of abuse can alter the levels of specific miRNAs that in turn result in dysregulation of target genes ultimately producing abnormal cellular function. The consequences of ethanol perturbation depends on the system under investigation, but may 1) lead to changes in populations of ion channel isoforms resulting in tolerance to the drug, 2) alter the fate of neuronal stem cells that may underlie the developmental abnormalities observed in FAS, 3) be involved in ethanol-induced GI conditions such as ALD and gut leakiness, and 4) alter inflammatory response that may ultimately impair wound and bone fracture healing. These findings demonstrate a striking diversity in the regulatory potential of miRNAs and underscore the importance of further investigations in the field.
Acknowledgements
The research reported here was supported by grants from NIH/NIAAA, #AA13440 to RCM, #AA017481 to AP, and #AA13745 to AK.
Footnotes
The following critical review condenses the proceedings of a symposium held at the 32nd Annual Meeting of the Research Society on Alcoholism, June 20–24, San Diego, CA.
miRNA annotation throughout this review is based on Human Genome Organization nomenclature (http://www.hugo-international.org/)
Non-union is due to inability of bony trabecular bridges to fuse the fractured ends, resulting in an inability of the fracture to heal.
References
- Adachi J, Mizoi Y, Fukunaga T, Ogawa Y, Ueno Y, Imamichi H. Degrees of alcohol intoxication in 117 hospitalized cases. J Stud Alcohol. 1991;52(5):448–453. doi: 10.15288/jsa.1991.52.448. [DOI] [PubMed] [Google Scholar]
- Adachi Y, Moore LE, Bradford BU, Gao W, Thurman RG. Antibiotics prevent liver injury in rats following long-term exposure to ethanol. Gastroenterology. 1995;108(1):218–24. doi: 10.1016/0016-5085(95)90027-6. [DOI] [PubMed] [Google Scholar]
- Akamatsu W, Okano HJ, Osumi N, Inoue T, Nakamura S, Sakakibara S, Miura M, Matsuo N, Darnell RB, Okano H. Mammalian ELAV-like neuronal RNA-binding proteins HuB and HuC promote neuronal development in both the central and the peripheral nervous systems. Proc Natl Acad Sci U S A. 1999;96(17):9885–90. doi: 10.1073/pnas.96.17.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113(6):673–6. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- Androulidaki A, Iliopoulos D, Arranz A, Doxaki C, Schworer S, Zacharioudaki V, Margioris AN, Tsichlis PN, Tsatsanis C. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. 2009;31(2):220–31. doi: 10.1016/j.immuni.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony TE, Klein C, Fishell G, Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. neuron. 2004;41(6):881–90. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- Atkinson NS, Robertson GA, Ganetzky B. A component of calcium-activated potassium channels encoded by the Drosophila slo locus. Science. 1991;253(5019):551–5. doi: 10.1126/science.1857984. [DOI] [PubMed] [Google Scholar]
- Barbato C, Ruberti F, Cogoni C. Searching for MIND: microRNAs in neurodegenerative diseases. J Biomed Biotechnol. 2009;2009:871313. doi: 10.1155/2009/871313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes DE, Walker DW. Prenatal ethanol exposure permanently reduces the number of pyramidal neurons in rat hippocampus. Brain Res. 1981;227(3):333–40. doi: 10.1016/0165-3806(81)90071-7. [DOI] [PubMed] [Google Scholar]
- Basak O, Taylor V. Identification of self-replicating multipotent progenitors in the embryonic nervous system by high Notch activity and Hes5 expression. Eur J Neurosci. 2007;25(4):1006–22. doi: 10.1111/j.1460-9568.2007.05370.x. [DOI] [PubMed] [Google Scholar]
- Bayer S, Altman J, Russo R, Zhang X. Timetables of neurogenesis in the human brain based on experimentally determined patterns in the rat. Neuro Toxicology. 1993;14(1):83–144. [PubMed] [Google Scholar]
- Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, Tamassia N, Mantovani A, Cassatella MA, Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc.Natl.Acad.Sci. 2009;106(13):5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand J, Floyd LL, Weber MK. Guidelines for identifying and referring persons with fetal alcohol syndrome. MMWR Recomm Rep. 2005;54(RR-11):1–14. [PubMed] [Google Scholar]
- Bhagwandeen BS, Apte M, Manwarring L, Dickeson J. Endotoxin induced hepatic necrosis in rats on an alcohol diet. J Pathol. 1987;152(1):47–53. doi: 10.1002/path.1711520107. [DOI] [PubMed] [Google Scholar]
- Bielawski DM, Zaher FM, Svinarich DM, Abel EL. Paternal alcohol exposure affects sperm cytosine methyltransferase messenger RNA levels. Alcohol Clin Exp Res. 2002;26(3):347–51. [PubMed] [Google Scholar]
- Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR. RNA editing of human microRNAs. Genome Biol. 2006;7(4):R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode C, Kugler V, Bode JC. Endotoxemia in patients with alcoholic and non-alcoholic cirrhosis and in subjects with no evidence of chronic liver disease following acute alcohol excess. J Hepatol. 1987;4(1):8–14. doi: 10.1016/s0168-8278(87)80003-x. [DOI] [PubMed] [Google Scholar]
- Braconi C, Patel T. MicroRNA expression profiling: a molecular tool for defining the phenotype of hepatocellular tumors. Hepatology. 2008;47(6):1807–9. doi: 10.1002/hep.22326. [DOI] [PubMed] [Google Scholar]
- Burbige EJ, Lewis DR, Jr., Halsted CH. Alcohol and the gastrointestinal tract. Med Clin North Am. 1984;68(1):77–89. doi: 10.1016/s0025-7125(16)31242-1. [DOI] [PubMed] [Google Scholar]
- Camarillo C, Miranda RC. Ethanol exposure during neurogenesis induces persistent effects on neural maturation: evidence from an ex vivo model of fetal cerebral cortical neuroepithelial progenitor maturation. Gene Expression. 2008 In Press. [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs Modulate Hematopoietic Lineage Differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Choudhury M, Shukla SD. Surrogate alcohols and their metabolites modify histone H3 acetylation: involvement of histone acetyl transferase and histone deacetylase. Alcohol Clin Exp Res. 2008;32(5):829–39. doi: 10.1111/j.1530-0277.2008.00630.x. [DOI] [PubMed] [Google Scholar]
- Clarren SK. Neuropathology in Fetal Alcohol Syndrome. In: West JR, editor. Alcohol and Brain Development. Oxford University Press; New York: 1986. pp. 158–166. [Google Scholar]
- Clarren SK, Alvord EC, Jr., Sumi SM, Streissguth AP, Smith DW. Brain malformations related to prenatal exposure to ethanol. J Pediatr. 1978;92(1):64–7. doi: 10.1016/s0022-3476(78)80072-9. [DOI] [PubMed] [Google Scholar]
- Clayburgh DR, Shen L, Turner JR. A porous defense: the leaky epithelial barrier in intestinal disease. Lab Invest. 2004;84(3):282–91. doi: 10.1038/labinvest.3700050. [DOI] [PubMed] [Google Scholar]
- Dolganiuc A, Petrasek J, Kodys K, Catalano D, Mandrekar P, Velayudham A, Szabo G. MicroRNA Expression Profile in Lieber-DeCarli Diet-Induced Alcoholic and Methionine Choline Deficient Diet-Induced Nonalcoholic Steatohepatitis Models in Mice. Alcohol Clin Exp Res. 2009 doi: 10.1111/j.1530-0277.2009.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworetzky SI, Trojnacki JT, Gribkoff VK. Cloning and expression of a human large-conductance calcium-activated potassium channel. Brain Res Mol Brain Res. 1994;27(1):189–93. doi: 10.1016/0169-328x(94)90203-8. [DOI] [PubMed] [Google Scholar]
- Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A Minicircuitry Comprised of MicroRNA-223 and Transcription Factors NFI-A and C/EBP-alpha Regulates Human Granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Ferguson SM, Brasnjo G, Hayashi M, Wolfel M, Collesi C, Giovedi S, Raimondi A, Gong LW, Ariel P, Paradise S, O'Toole E, Flavell R, Cremona O, Miesenbock G, Ryan TA, De Camilli P. A selective activity-dependent requirement for dynamin 1 in synaptic vesicle endocytosis. Science. 2007;316(5824):570–4. doi: 10.1126/science.1140621. [DOI] [PubMed] [Google Scholar]
- Flatscher-Bader T, van der Brug M, Hwang JW, Gochee PA, Matsumoto I, Niwa S, Wilce PA. Alcohol-responsive genes in the frontal cortex and nucleus accumbens of human alcoholics. J Neurochem. 2005;93(2):359–70. doi: 10.1111/j.1471-4159.2004.03021.x. [DOI] [PubMed] [Google Scholar]
- Foulk DA, Szabo RM. Diaphyseal humerus fractures: Natural history and occurance of nonunion. Orthopedics. 1995;18(4):333–335. doi: 10.3928/0147-7447-19950401-04. [DOI] [PubMed] [Google Scholar]
- Galambos JT. Alcoholic hepatitis: its therapy and prognosis. Prog Liver Dis. 1972;4:567–88. [PubMed] [Google Scholar]
- Garro AJ, McBeth DL, Lima V, Lieber CS. Ethanol consumption inhibits fetal DNA methylation in mice: implications for the fetal alcohol syndrome. Alcohol Clin Exp Res. 1991;15(3):395–8. doi: 10.1111/j.1530-0277.1991.tb00536.x. [DOI] [PubMed] [Google Scholar]
- Gotz M, Barde YA. Radial glial cells defined and major intermediates between embryonic stem cells and CNS neurons. neuron. 2005;46(3):369–72. doi: 10.1016/j.neuron.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Grant BF, Dufour MC, Harford TC. Epidemiology of alcoholic liver disease. Semin Liver Dis. 1988;8(1):12–25. doi: 10.1055/s-2008-1040525. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid A, Wani NA, Kaur J. New perspectives on folate transport in relation to alcoholism-induced folate malabsorption--association with epigenome stability and cancer development. FEBS J. 2009;276(8):2175–91. doi: 10.1111/j.1742-4658.2009.06959.x. [DOI] [PubMed] [Google Scholar]
- Harper CG, Kril JJ, Holloway RL. Brain shrinkage in chronic alcoholics: a pathological study. Br Med J (Clin Res Ed) 1985;290(6467):501–4. doi: 10.1136/bmj.290.6467.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin D. Classification of alcohol use disorders. Alcohol Res Health. 2003;27(1):5–17. [PMC free article] [PubMed] [Google Scholar]
- Hebert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32(4):199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Hebert SS, Horre K, Nicolai L, Bergmans B, Papadopoulou AS, Delacourte A, De Strooper B. MicroRNA regulation of Alzheimer's Amyloid precursor protein expression. Neurobiol Dis. 2009;33(3):422–8. doi: 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Kerin MJ. MicroRNAs as Novel Biomarkers for Breast Cancer. J Oncol. 2009;2009:950201. doi: 10.1155/2010/950201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino K, Tsuchiya K, Fukao T, Kiga K, Okamoto R, Kanai T, Watanabe M. Inducible expression of microRNA-194 is regulated by HNF-1alpha during intestinal epithelial cell differentiation. Rna. 2008;14(7):1433–42. doi: 10.1261/rna.810208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander D. The intestinal permeability barrier. A hypothesis as to its regulation and involvement in Crohn's disease. Scand J Gastroenterol. 1992;27(9):721–6. doi: 10.3109/00365529209011172. [DOI] [PubMed] [Google Scholar]
- Huang W, Li MD. Nicotine modulates expression of miR-140*, which targets the 3'-untranslated region of dynamin 1 gene (Dnm1) Int J Neuropsychopharmacol. 2009;12(4):537–46. doi: 10.1017/S1461145708009528. [DOI] [PubMed] [Google Scholar]
- Ivanov AI, Nusrat A, Parkos CA. Endocytosis of the apical junctional complex: mechanisms and possible roles in regulation of epithelial barriers. Bioessays. 2005;27(4):356–65. doi: 10.1002/bies.20203. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Yamamoto M, Ozawa H, Saito T, Kato T. Decreased expression of NEFH and PCP4/PEP19 in the prefrontal cortex of alcoholics. Neurosci Res. 2004;49(4):379–85. doi: 10.1016/j.neures.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451(7182):1125–9. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A. 2009;106(31):13052–7. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalant H. Research on tolerance: what can we learn from history? Alcohol Clin Exp Res. 1998;22(1):67–76. doi: 10.1111/j.1530-0277.1998.tb03618.x. [DOI] [PubMed] [Google Scholar]
- Kapeller J, Houghton LA, Monnikes H, Walstab J, Moller D, Bonisch H, Burwinkel B, Autschbach F, Funke B, Lasitschka F, Gassler N, Fischer C, Whorwell PJ, Atkinson W, Fell C, Buchner KJ, Schmidtmann M, van der Voort I, Wisser AS, Berg T, Rappold G, Niesler B. First evidence for an association of a functional variant in the microRNA-510 target site of the serotonin receptor-type 3E gene with diarrhea predominant irritable bowel syndrome. Hum Mol Genet. 2008;17(19):2967–77. doi: 10.1093/hmg/ddn195. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavarzian A, Choudhary S, Holmes EW, Yong S, Banan A, Jakate S, Fields JZ. Preventing gut leakiness by oats supplementation ameliorates alcohol-induced liver damage in rats. J Pharmacol Exp Ther. 2001;299(2):442–8. [PubMed] [Google Scholar]
- Keshavarzian A, Farhadi A, Forsyth CB, Rangan J, Jakate S, Shaikh M, Banan A, Fields JZ. Evidence that chronic alcohol exposure promotes intestinal oxidative stress, intestinal hyperpermeability and endotoxemia prior to development of alcoholic steatohepatitis in rats. J Hepatol. 2009;50(3):538–47. doi: 10.1016/j.jhep.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavarzian A, Holmes EW, Patel M, Iber F, Fields JZ, Pethkar S. Leaky gut in alcoholic cirrhosis: a possible mechanism for alcohol-induced liver damage. Am J Gastroenterol. 1999;94(1):200–7. doi: 10.1111/j.1572-0241.1999.00797.x. [DOI] [PubMed] [Google Scholar]
- Klatsky AL. Alcohol and cardiovascular diseases. Expert Rev Cardiovasc Ther. 2009;7(5):499–506. doi: 10.1586/erc.09.22. [DOI] [PubMed] [Google Scholar]
- Komatsu S, Sakata-Haga H, Sawada K, Hisano S, Fukui Y. Prenatal exposure to ethanol induces leptomeningeal heterotopia in the cerebral cortex of the rat fetus. Acta Neuropathol (Berl) 2001;101:22–26. doi: 10.1007/s004010000257. [DOI] [PubMed] [Google Scholar]
- Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, Mendell JR, Mendell JT. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137(6):1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotkoskie LA, Norton S. Prenatal brain malformations following acute ethanol exposure in the rat. Alcohol Clin Exp Res. 1988;12(6):831–836. doi: 10.1111/j.1530-0277.1988.tb01355.x. [DOI] [PubMed] [Google Scholar]
- Kotkoskie LA, Norton S. Morphometric analysis of developing rat cerebral cortex following acute prenatal ethanol exposure. 1989;106(3):283–288. doi: 10.1016/0014-4886(89)90161-1. [DOI] [PubMed] [Google Scholar]
- Kotzot D. Maternal uniparental disomy 7 and Silver-Russell syndrome - clinical update and comparison with other subgroups. Eur J Med Genet. 2008;51(5):444–51. doi: 10.1016/j.ejmg.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Kril JJ, Halliday GM, Svoboda MD, Cartwright H. The cerebral cortex is damaged in chronic alcoholics. Neuroscience. 1997;79(4):983–98. doi: 10.1016/s0306-4522(97)00083-3. [DOI] [PubMed] [Google Scholar]
- Kril JJ, Harper CG. Neuronal counts from four cortical regions of alcoholic brains. Acta Neuropathol (Berl) 1989;79(2):200–4. doi: 10.1007/BF00294379. [DOI] [PubMed] [Google Scholar]
- Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci. 2008;28(45):11720–30. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laukoetter MG, Bruewer M, Nusrat A. Regulation of the intestinal epithelial barrier by the apical junctional complex. Curr Opin Gastroenterol. 2006;22(2):85–9. doi: 10.1097/01.mog.0000203864.48255.4f. [DOI] [PubMed] [Google Scholar]
- Lefebvre L, Viville S, Barton SC, Ishino F, Keverne EB, Surani MA. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat Genet. 1998;20(2):163–169. doi: 10.1038/2464. [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA. Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcohol Clin Exp Res. 2000;24(12):1873–82. [PubMed] [Google Scholar]
- Li Y, Wang F, Lee J-A, Gao F-B. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes & Development. 2006;20(20):2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Dodd PR, Randall PK, Harris RA, Mayfield RD. Gene expression profiling of individual cases reveals consistent transcriptional changes in alcoholic human brain. J Neurochem. 2004;90(5):1050–8. doi: 10.1111/j.1471-4159.2004.02570.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, Mayfield RD. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2006;31(7):1574–82. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- Maher JJ. Alcoholic steatosis and steatohepatitis. Semin Gastrointest Dis. 2002;13(1):31–9. [PubMed] [Google Scholar]
- Mayfield RD, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J Neurochem. 2002;81(4):802–13. doi: 10.1046/j.1471-4159.2002.00860.x. [DOI] [PubMed] [Google Scholar]
- Michael MZ, SM OC, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1(12):882–91. [PubMed] [Google Scholar]
- Miller M. Effects of prenatal exposure to ethanol on neocortical development: II. Cell proliferation in the ventricular and subventricular zones of the rat. The Journal of Comparative Neurology. 1989;287:326–338. doi: 10.1002/cne.902870305. [DOI] [PubMed] [Google Scholar]
- Miller MW. Limited ethanol exposure selectively alters the proliferation of precursor cells in the cerebral cortex. Alcohol Clin Exp Res. 1996;20(1):139–43. doi: 10.1111/j.1530-0277.1996.tb01056.x. [DOI] [PubMed] [Google Scholar]
- Miller MW, Nowakowski RS. Effect of prenatal exposure to ethanol on the cell cycle kinetics and growth fraction in the proliferative zones of fetal rat cerebral cortex. Alcohol Clin Exp Res. 1991;15(2):229–32. doi: 10.1111/j.1530-0277.1991.tb01861.x. [DOI] [PubMed] [Google Scholar]
- Montag J, Hitt R, Opitz L, Schulz-Schaeffer WJ, Hunsmann G, Motzkus D. Upregulation of miRNA hsa-miR-342-3p in experimental and idiopathic prion disease. Mol Neurodegener. 2009;4(1):36. doi: 10.1186/1750-1326-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney SM, Siegenthaler JA, Miller MW. Ethanol Induces Heterotopias in Organotypic Cultures of Rat Cerebral Cortex. Cereb Cortex. 2004;14(10):1071–80. doi: 10.1093/cercor/bhh066. [DOI] [PubMed] [Google Scholar]
- Nanji AA, Khettry U, Sadrzadeh SM. Lactobacillus feeding reduces endotoxemia and severity of experimental alcoholic liver (disease) Proc Soc Exp Biol Med. 1994;205(3):243–7. doi: 10.3181/00379727-205-43703. [DOI] [PubMed] [Google Scholar]
- Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of colorectal cancer patients: A potential marker for colorectal cancer screening. Gut. 2009a doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- Ng EK, Wong CL, Ma ES, Kwong A. MicroRNAs as New Players for Diagnosis, Prognosis, and Therapeutic Targets in Breast Cancer. J Oncol. 2009b;2009:305420. doi: 10.1155/2009/305420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyquist F, Berglund M, Nilsson BE, Obrant KJ. Nature and Healing of Tibial Shaft Fractures in Alcohol Abusers. Alcohol & Alcoholism. 1997;32(1):91–95. doi: 10.1093/oxfordjournals.alcalc.a008240. [DOI] [PubMed] [Google Scholar]
- Nyquist F, Overgaard A, DÅppe H, Obrant KJ. Alcohol Abuse and Healing Complications After Cervical Hip Fractures. Alcohol & Alcoholism. 1998;33(4):373–380. doi: 10.1093/oxfordjournals.alcalc.a008407. [DOI] [PubMed] [Google Scholar]
- O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hemaopoietic stem cells causes a myeloproliferative disorder. J.Exp.Med. 2008;205(3):585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc.Natl.Acad.Sci. 2007;104(5):1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouko LA, Shantikumar K, Knezovich J, Haycock P, Schnugh DJ, Ramsay M. Effect of alcohol consumption on CpG methylation in the differentially methylated regions of H19 and IG-DMR in male gametes: implications for fetal alcohol spectrum disorders. Alcohol Clin Exp Res. 2009;33(9):1615–27. doi: 10.1111/j.1530-0277.2009.00993.x. [DOI] [PubMed] [Google Scholar]
- Pietrzykowski A, Treistman S. The molecular basis of tolerance. Alcohol Res Health. 2008;31(4):298–309. [PMC free article] [PubMed] [Google Scholar]
- Pietrzykowski AZ, Friesen RM, Martin GE, Puig SI, Nowak CL, Wynne PM, Siegelmann HT, Treistman SN. Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron. 2008;59(2):274–87. doi: 10.1016/j.neuron.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prock TL, Miranda RC. Embryonic cerebral cortical progenitors are resistant to apoptosis, but increase expression of suicide receptor DISC-complex genes and suppress autophagy following ethanol exposure. Alcohol Clin Exp Res. 2007;31(4):694–703. doi: 10.1111/j.1530-0277.2007.00354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi JJ. New hope for a microRNA therapy for liver cancer. Cell. 2009;137(6):990–2. doi: 10.1016/j.cell.2009.05.038. [DOI] [PubMed] [Google Scholar]
- Salero E, Hatten ME. Differentiation of ES cells into cerebellar neurons. Proc Natl Acad Sci U S A. 2007;104(8):2997–3002. doi: 10.1073/pnas.0610879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom SN, Hebert JM, Thammongkol U, Smith J, Nisbet G, Surani MA, McConnell SK, Livesey FJ. Genomic characterisation of a Fgf-regulated gradient-based neocortical protomap. Development. 2005;132(17):3947–61. doi: 10.1242/dev.01968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santillano DR, Kumar LS, Prock TL, Camarillo C, Tingling JD, Miranda RC. Ethanol induces cell-cycle activity and reduces stem cell diversity to alter both regenerative capacity and differentiation potential of cerebral cortical neuroepithelial precursors. BMC Neurosci. 2005;6:59. doi: 10.1186/1471-2202-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyan P, Golden HB, Miranda RC. Competing interactions between microRNAs determine neural progenitor survival and proliferation following ethanol exposure: Evidence from an ex vivo model of the fetal cerebral cortical neuroepithelium. J Neurosci. 2007;27(32):8546–57. doi: 10.1523/JNEUROSCI.1269-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan H, Zhang Y, Lu Y, Pan Z, Cai B, Wang N, Li X, Feng T, Hong Y, Yang B. Downregulation of miR-133 and miR-590 contributes to nicotine-induced atrial remodelling in canines. Cardiovasc Res. 2009;83(3):465–72. doi: 10.1093/cvr/cvp130. [DOI] [PubMed] [Google Scholar]
- Shibata M, Kurokawa D, Nakao H, Ohmura T, Aizawa S. MicroRNA-9 Modulates Cajal-Retzius Cell Differentiation by Suppressing Foxg1 Expression in Mouse Medial Pallium. J. Neurosci. 2008;28(41):10415–10421. doi: 10.1523/JNEUROSCI.3219-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SD, Velazquez J, French SW, Lu SC, Ticku MK, Zakhari S. Emerging role of epigenetics in the actions of alcohol. Alcohol Clin Exp Res. 2008;32(9):1525–34. doi: 10.1111/j.1530-0277.2008.00729.x. [DOI] [PubMed] [Google Scholar]
- Sokol RJ, Delaney-Black V, Nordstrom B. Fetal alcohol spectrum disorder. JAMA. 2003;290(22):2996–9. doi: 10.1001/jama.290.22.2996. [DOI] [PubMed] [Google Scholar]
- Sokolov BP, Jiang L, Trivedi NS, Aston C. Transcription profiling reveals mitochondrial, ubiquitin and signaling systems abnormalities in postmortem brains from subjects with a history of alcohol abuse or dependence. J Neurosci Res. 2003;72(6):756–67. doi: 10.1002/jnr.10631. [DOI] [PubMed] [Google Scholar]
- Szabo G. Consequences of alcohol consumption on host defence. Alcohol Alcohol. 1999;34(6):830–41. doi: 10.1093/alcalc/34.6.830. [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF- k B-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc.Natl.Acad.Sci. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Banan A, Forsyth CB, Fields JZ, Lau CK, Zhang LJ, Keshavarzian A. Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcohol Clin Exp Res. 2008;32(2):355–64. doi: 10.1111/j.1530-0277.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- Tang Y, Forsyth CB, Banan A, Fields JZ, Keshavarzian A. Oats supplementation prevents alcohol-induced gut leakiness in rats by preventing alcohol-induced oxidative tissue damage. J Pharmacol Exp Ther. 2009a;329(3):952–8. doi: 10.1124/jpet.108.148643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Forsyth CB, Farhadi A, Rangan J, Jakate S, Shaikh M, Banan A, Fields JZ, Keshavarzian A. Nitric Oxide-Mediated Intestinal Injury Is Required for Alcohol-Induced Gut Leakiness and Liver Damage. Alcohol Clin Exp Res. 2009b doi: 10.1111/j.1530-0277.2009.00946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tili E, Michaille JJ, Cimmino A, Costinean S, Dumitru GA, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM. Modulation of miR-155 and miR-125b Levels following Lipopolysccharide/TNF-a Stimulation and Their Possible Roles in Regulating the Response to Endotoxin Shock. J.Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- Tonnesen H, Pedersen A, Jensen MR, Moller A, Madsen JC. Ankle Fractures and Alcoholism. The Influence of Alcoholism on Morbidity after Malleolar Fractures. J.Bone Joint Surg. 1991;73-B:511–513. doi: 10.1302/0301-620X.73B3.1670461. [DOI] [PubMed] [Google Scholar]
- Tryndyak VP, Ross SA, Beland FA, Pogribny IP. Down-regulation of the microRNAs miR-34a, miR-127, and miR-200b in rat liver during hepatocarcinogenesis induced by a methyl-deficient diet. Mol Carcinog. 2009;48(6):479–87. doi: 10.1002/mc.20484. [DOI] [PubMed] [Google Scholar]
- Turner JR. Molecular basis of epithelial barrier regulation: from basic mechanisms to clinical application. Am J Pathol. 2006;169(6):1901–9. doi: 10.2353/ajpath.2006.060681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangipuram SD, Grever WE, Parker GC, Lyman WD. Ethanol increases fetal human neurosphere size and alters adhesion molecule gene expression. Alcohol Clin Exp Res. 2008;32(2):339–47. doi: 10.1111/j.1530-0277.2007.00568.x. [DOI] [PubMed] [Google Scholar]
- Verrier JD, Lau P, Hudson L, Murashov AK, Renne R, Notterpek L. Peripheral myelin protein 22 is regulated post-transcriptionally by miRNA-29a. Glia. 2009;57(12):1265–79. doi: 10.1002/glia.20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LL, Zhang Z, Li Q, Yang R, Pei X, Xu Y, Wang J, Zhou SF, Li Y. Ethanol exposure induces differential microRNA and target gene expression and teratogenic effects which can be suppressed by folic acid supplementation. Hum Reprod. 2009a;24(3):562–79. doi: 10.1093/humrep/den439. [DOI] [PubMed] [Google Scholar]
- Wang QZ, Xu W, Habib N, Xu R. Potential uses of microRNA in lung cancer diagnosis, prognosis, and therapy. Curr Cancer Drug Targets. 2009b;9(4):572–94. doi: 10.2174/156800909788486731. [DOI] [PubMed] [Google Scholar]
- Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008a;135(5):1624–1635. e24. doi: 10.1053/j.gastro.2008.07.068. [DOI] [PubMed] [Google Scholar]
- Wu Q, Law PY, Wei LN, Loh HH. Post-transcriptional regulation of mouse mu opioid receptor (MOR1) via its 3' untranslated region: a role for microRNA23b. FASEB J. 2008b;22(12):4085–95. doi: 10.1096/fj.08-108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Zhang L, Law PY, Wei LN, Loh HH. Long-term morphine treatment decreases the association of mu-opioid receptor (MOR1) mRNA with polysomes through miRNA23b. Mol Pharmacol. 2009;75(4):744–50. doi: 10.1124/mol.108.053462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Liao X, Wong C. Down-regulations of B-cell lymphoma 2 (Bcl-2) and myeloid cell leukemia sequence 1 (Mcl-1) by MicroRNA 153 induce apoptosis in a glioblastoma cell line DBTRG-05MG. Int J Cancer. 2009 doi: 10.1002/ijc.24823. [DOI] [PubMed] [Google Scholar]
- Yang L, Belaguli N, Berger DH. MicroRNA and colorectal cancer. World J Surg. 2009;33(4):638–46. doi: 10.1007/s00268-008-9865-5. [DOI] [PubMed] [Google Scholar]
- Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16(4):365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]