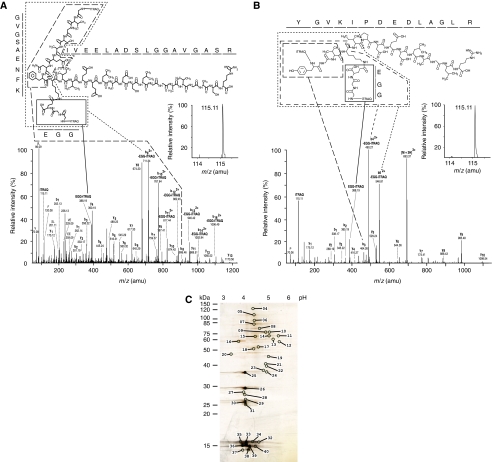
Figure 2.
Specific enrichment of pupylated peptide in polyhistidine affinity captured eluate. (A, B) Annotated CID spectra that contributed to the identification of (A) MSMEG_2352 and (B) MSMEG_4326 as a pupylation targets. The CID spectrum of (A) was derived by fragmentation of a quadruple-charged precursor ion observed at m/z [766.91+4H]4+ and was matched to the branched peptide GVGSAENFK(QGG)IVEELADSLGGAVGASR carrying iTRAQ labels at the N-termini of both its main and branched chain. Please note the absence of a detectable iTRAQ114 signature mass peak (inset), indicating that this peptide was exclusively contributed by the pupylated iTRAQ 115-labeled sample. Also note the double charged nature of branched peptide fragments due to the retention of an additional charge by the primary amine present within the N-terminal glycine of the GGQ pupylation stub. The CID spectrum of (B) was derived by fragmentation of a triple charged precursor ion observed at m/z [693.37 +3H]3+ and was matched to the branched peptide YGVK(QGG)IPDEDLAGLR, carrying iTRAQ labels at the N-termini of both its main and branched chain. For information on additional peptides see Table I. (C) Complementary 2D gel analysis. Pupylated substrates were separated on a 2D gel and spots are numbered, which are subjected to tandem MS/MS analyses. The numbering of the spots corresponds to the numbering used in the tables. The pH range of the first dimension and apparent molecular weight of second dimension are denoted.