Figure 1.
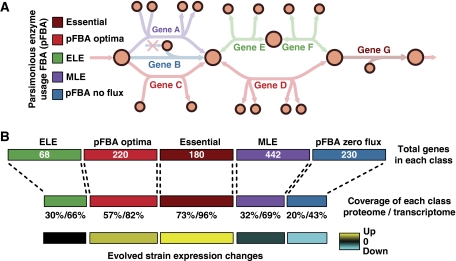
A variant of flux balance analysis shows consistency with proteomic and transcriptomic data. Parsimonious enzyme usage FBA (pFBA) is used to label all metabolic genes based on simulation results. (A) pFBA classifies each gene based on its ability to contribute to the optimal growth rate predictions and its flux level. These classes include (1) essential genes; (2) pFBA optima, which includes genes that are predicted to be used for optimal growth in silico; (3) ELE, which includes genes that will increase cellular metabolic flux if used; (4) MLE, which includes genes predicted to decrease the growth rate if used; and (5) pFBA no-flux, which includes genes that cannot be used in the given growth conditions. (B) The omic data show good coverage of essential genes and the pFBA optima, and low coverage of the genes that are predicted to be non-functional. In addition, in laboratory evolution experiments, genes within these optimal states are upregulated, whereas non-functional genes are downregulated. These results support predicted optimal growth states, and suggest that laboratory-evolved strains further enhance these optimal growth states.