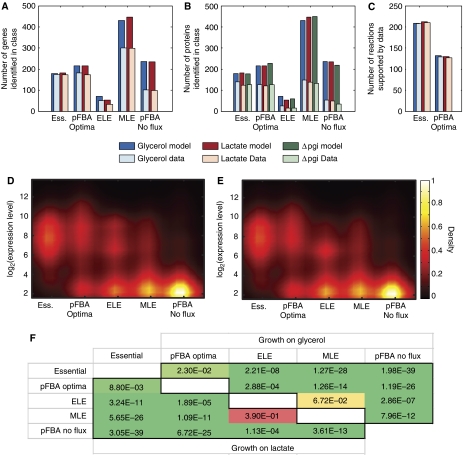
Figure 3.
pFBA classes are consistent with omic data. Simulations for each growth condition were used to classify each gene according to the efficiency of its associated reaction(s). The coverage of (A) expressed genes above a statistical cutoff and (B) identified proteins were determined. More genes and proteins in the essential and pFBA optima classes were expressed than genes and proteins in the less efficient (ELE and MLE) and conditionally non-functional (pFBA no-flux) pathways. Almost half of the missing essential and pFBA optima proteins are membrane associated, hence its lower coverage. (C) In all, 99 and 98% of the active reactions associated with the essential genes and pFBA optima, respectively, are supported by the union of expressed genes and proteins. Gene expression levels are also consistent with the pFBA classes, as shown in density plots for growth on (D) glycerol and (E) lactate. (F) Pairwise comparisons of classes show the significant ordering of expression levels as Essential>pFBA optima>ELE>MLE>pFBA no-flux for growth on glycerol (upper triangle) and lactate (lower triangle) (one-sided Wilcoxon test).