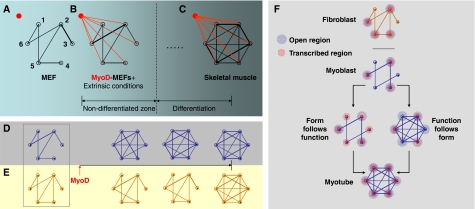
Figure 1.
Reprogramming: a network view. Network diagrams (A–C) are representations of collective information from chromosomal topology (D) and transcriptome (E) networks, that is the biological network, where nodes are genes or chromosomes. Thus, we define the network in (A) as a nuclear circuit, composed of the networks in (D, E) (rectangular outline). (A) A biological network representing the specific network signature of the MEF, where some genes are connected, and some are not. The connectivity is weighted, that is the strength of the connection depends on the gene pair, shown by edge thickness. For example, connectivity between genes 2 and 5 is weaker than between 2 and 3. MyoD (the red node) is part of the network but in this case has no connection with other genes as it not expressed. (B) Activated MyoD establishes connections with the rest of the genes and initiates rewiring, accompanied by an increase in MyoD binding affinity. (C) The differentiated system has unique network architecture; new edges appear, resulting in a new network. Note that between (B) and (C) other network structures exist but we have only shown a few points during the process. (D, E) (Within-network organization): the MEF network initially exists in a unique pattern before induction of MyoD. After MyoD induction, the chromosomal network begins to rewire, increasing its connectivity and indicating changes in spatial architecture. The transcriptome network lags behind but gradually increases connectivity, and reaches a network state similar to the chromosomal network (higher communication). (F) Schematic representation of the question: is form a precondition for, or does form follow, function? The fibroblast and myoblast differ in nuclear architecture. In both cell types, actively transcribed genes (red circles) will be localized in active regions of chromatin (blue circles) and show a network of physical adjacencies (edges) within other regions of active chromatin. If form follows function, then muscle gene expression will precede or coincide with localization of a gene to an active region and rearrangement of nuclear adjacencies; whereas if function follows form, then repositioning of genes to active regions and rearrangement of nuclear adjacencies will precede gene transcription.