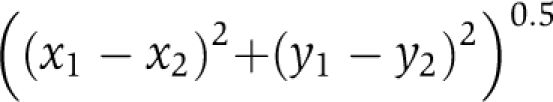|
Adjacency matrix of a graph: Square matrix with aij=1, when there is an edge from node i to node j; otherwise aij=0. Weighted adjacency matrices with entries that are not simply zero or 1 have entries equal instead to the weights on the edges |
|
Attractor: An attractor is the end-state of a dynamic system as it moves over time. Attractors may be fixed points, periodic, or chaotic and may also be stable or unstable |
|
Basin of attraction: A region in phase space associated with a given attractor. The basin of attraction of an attractor is the set of all (initial) points that move toward that attractor |
|
Cellular differentiation: The dynamical process of a less specialized cell transitioning into a more specialized cell type |
|
Cellular reprogramming: The process of changing a mature unipotent adult cell's unique genetic and epigenetic signature, typically by manipulating signal transduction mechanisms and growth factors, so as to confer plasticity, pluripotency, or ability to differentiate into at least one other type of cell |
|
Chromosomal interaction network: Nodes are chromosome or genes and the edges are computed based on proximity of chromosomes. Spatial relationships between each pair of chromosomes include distance between centroids, closest distance, and also more complex relationships such as shared volume and contact area |
| |
|
Communication: Interaction between networks. We quantify network communication by comparing the similarity between the network's corresponding weighted adjacency matrices |
|
Connectivity: Interaction within a network. We quantify network connectivity using the second smallest eigenvalue of the network's Laplacian matrix |
| |
|
Dynamical system: An evolution rule that defines a trajectory as a function of a single parameter (time) on a set of states (the phase space) is a dynamical system |
| |
|
E-box: Is a regulatory DNA sequence that usually lies upstream of a gene in a promoter region, commonly bound by basic helix-loop-helix transcription factors |
|
Eigenvalues: A special set of scalars associated with linear systems of equations. They are also known as characteristic roots. The decomposition of a square matrix A into eigenvalues and eigenvectors (the vectors associated with eigenvalues) is known in this work as eigen decomposition. Eigenvalues are represented by λ and eigenvectors by x. Ax=λx with x≠0 so det(A–λI)=0 |
Euclidean distance: Is the ‘ordinary' distance between two points. In a plane with p1 at (x1,y1) and p2 at (x2,y2), it is 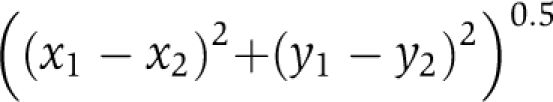
|
|
Graph G: A set of n nodes connected pairwise by m edges. In a complete graph, all 0.5(n(n−1)) edges between nodes |
|
Nonlinear: A function that is not linear. Most things in nature are nonlinear. This means that in a very real way, the whole is at least different from the sum of the parts |
|
Spectral karyotyping (SKY): A method of visualizing all chromosomes in the genome simultaneously, with each chromosome labeled with a unique color |
|
Symmetric matrix: A matrix with the lower-left half equal to the mirror image of the upper-right half |
|
Trace: Let A be an n × n matrix with eigenvalues λ1,λ2,…,λn. The sum of the eigenvalues is the sum of the diagonal entries of A and is called the trace of A
|
|
Transcriptome network: Nodes are chromosome or genes and the edges are computed based on gene coregulation. One of the measures of gene coregulation is the relative entropy between gene profiles |