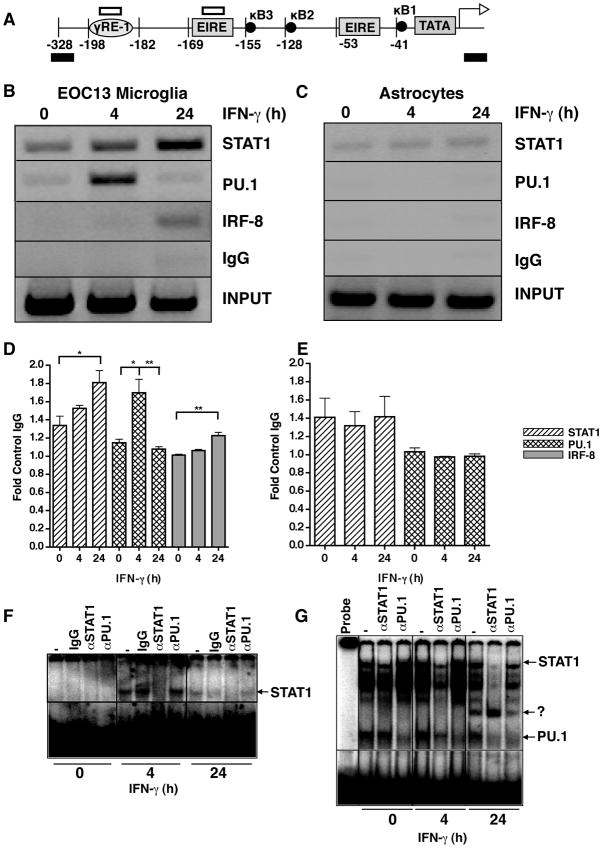
Figure 4.
Interaction of STAT1, PU.1 and IRF-8 with the Cxcl9 gene promoter in microglia and astrocytes. A schematic illustration of the murine Cxcl9 gene promoter (A) showing the location of the primer sites for ChIP analysis (black bars) and the sites corresponding to the oligonucleotide probe used for EMSA (open bars). ChIP analysis was performed on EOC microglia (B) or astrocytes (C). Cells were incubated in the absence or presence of IFN-γ (1000U/ml) for 0, 4 or 24 hours, and cross-linked with formaldehyde and soluble chromatin was subjected to immunoprecipitation with antibodies against STAT1, PU.1, IRF-8 or normal IgG as described in the Materials and Methods. Representative PCR images taken from 1 of 3 independent experiments is shown for EOC-13 cells (A) and astrocytes (B). Fold-change as compared with IgG was quantified for each transcription factor binding for EOC13 cells (D) and astrocytes (E). For statistical significance: *p<0.05, **p<0.01, ***p<0.001. Binding activity to γ-RE (F) or EIRE1 (G) radiolabeled oligonucleotides with nuclear extracts (5 μg) from EOC-13 cells treated with IFN-γ for the times shown and incubated with the indicated antibodies was performed as described in Materials and Methods.