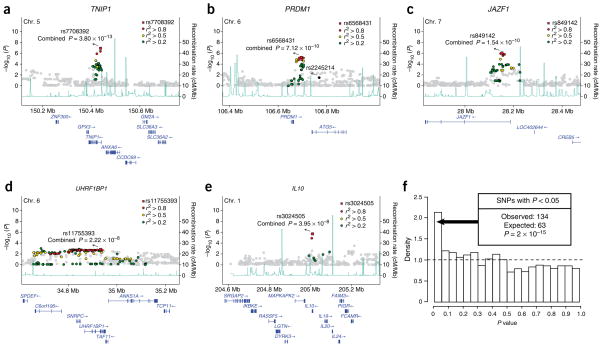
Figure 2.
Newly discovered genome-wide significant associations in SLE. (a–e) Association results from the GWA scan are plotted on the y axis versus genomic position on the indicated chromosome on the x axis within a 500-kb region surrounding the loci defined by (a) TNIP1, (b) PRDM1, (c) JAZF1, (d) UHRF1BP1 and (e) IL10. The meta-analysis P value for the most strongly associated marker is indicated by a red square. P values from the genome scan are shaded to indicate LD to the genome-wide associated variant: red, r2 > 0.8; yellow, r2 > 0.5; green, r2 > 0.2; gray, r2 < 0.2. Along the bottom are the recombination rates from the CEU HapMap (light blue line) and the known human genes (blue). A previously reported and independent SLE risk locus at the nearby ATG5 gene is indicated (b; rs2245214). (f) Histogram of P values of 1,256 independent SNPs (r2 < 0.1 to any other SNP in the array) in the 1,963 case and 4,329 control replication samples. Under a null distribution, the expected density of results is indicated by the dashed line. A significant enrichment of results with P < 0.05 was observed.