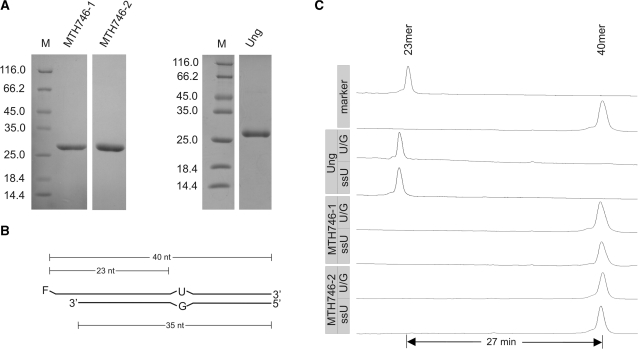
Figure 2.
Test for uracil DNA glycosylase activity associated with MTH746-1 and MTH746-2 purified from the Ung-deficient E. coli strain BL21_UX. (A) SDS–PAGE analysis (Coomassie stained) of purified proteins after IMAC and heparin affinity chromatography. MTH746 was N-terminally truncated by either omitting the first 19 amino acids (named MTH746-1) or the first 26 amino acids (named MTH746-2). For clarification, the first amino acid post each truncation is indicated in bold in the amino acid sequence of MTH746 depicted in Figure 1. For technical reasons, the N-terminal amino acid residues of MTH746-1 were changed from VF to MI. Relative molecular weights of marker proteins [×10−3] in lane M are indicated to the left. The gel was stained with Coomassie brilliant blue R-250. Ung, uracil-N-glycosylase from E. coli. Calculated relative molecular masses are 28 020 for MTH746-1, 27 040 for MTH746-2 and 26 760 for Ung. For production and purification details, see ‘Materials and Methods’ section. (B) Schematic drawing of the double-stranded U/G mismatched DNA substrate. F, fluorescein moeity. The lengths of the substrate oligonucleotides and the expected products are indicated. For the single-stranded DNA substrate (ssU) the fluorescently labeled oligonucleotide without the complementary strand was used. (C) Track recording of the ALF DNA sequencer after glycosylase assay with E. coli uracil-N-glycosylase Ung (control) or MTH746-1 and MTH746-2, respectively. Incubation was performed with 24 pmol of the respective protein and 0.12 pmol of substrates for 30 min at 37°C. Numbers of minutes refer to run-time differences between marker oligonucleotides. Note that the product in the Ung control is migrating faster than the 23-mer marker oligonucleotide, since it contains a phosphate residue at its 3′-terminus left behind from two rounds of elimination reaction with NaOH. For reaction details refer to ‘Materials and Methods’ section.