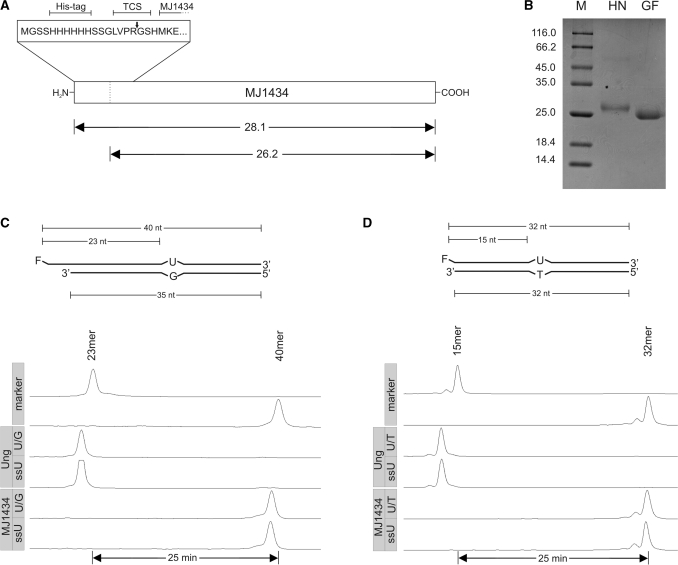
Figure 3.
Uracil DNA glycosylase assay with MJ1434. (A) Schematic drawing of the expression construct. TCS, thrombin cleavage site. The arrows indicate the expected relative molecular masses [×10−3] of MJ1434 before and after thrombin cleavage. (B) SDS–PAGE analysis of purified MJ1434 after heparin chromatography (HN) and after thrombin cleavage and gel filtration chromatography (GF). Labels as in Figure 2A. (C) Top: schematic drawing of the double-stranded U/G mismatched DNA substrate as in Figure 2B. Bottom: trackings of fluorescence readout as recorded by an ALF DNA sequencer after glycosylase assays with MJ1434 (5 pmol) using double-stranded (U/G) or single-stranded (ssU) uracil containing oligonucleotide substrates (1 pmol each). Ung was used as the positive control. Labels as in Figure 2C. (D) Top: scheme of U/T mismatched substrate. Bottom: readout of glycosylase assay with Ung (control) or MJ1434 as in (C) but with substrates bearing the identical sequence that was used by Chung et al. (23). For details refer to ‘Materials and Methods’ section.