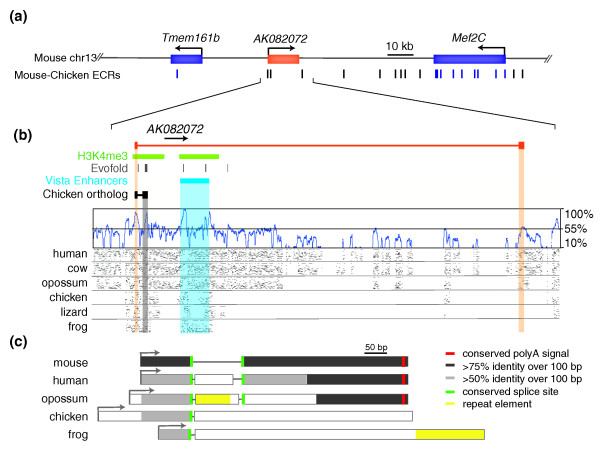
Figure 3.
Evolutionary constraint of AK082072. (a) The genomic region of mouse chromosome 13 (chr13) encompassing lncRNA AK082072 (523 bp) is depicted. Note the locations of the flanking protein-coding genes: Tmem161b (transmembrane protein 161b) and Mef2C (myocyte enhancer factor 2C). (b) A more detailed representation of AK082072 (exons highlighted in orange) and its immediate flanking regions. Below the gene structures are the positions of H3K4me3 chromatin marks (green) detected in mouse brain, VISTA conserved non-coding midbrain enhancer element 268 (obtained from the UCSC Genome Browser), and a BLAT alignment of the chicken AK082072 ortholog, as well as similar tracks as those in Figure 2b. Note the detected homology with orthologous frog sequence in exon 1. (c) Conservation and relative sizes of AK082072 orthologs in various species. Note the sequence conservation (relative to the mouse sequence) at both the 5' and 3' ends and the conserved position of splice sites (green). Unlike the other vertebrate genomes considered, the zebra finch genome did not align to the proximal promoter or first exon of mouse AK082072. This apparent lack of sequence identity might reflect either an unannotated gap in its genome assembly or rapidly evolving sequence within its orthologous genomic region. Other details are provided in the legend to Figure 2. ECR, evolutionarily conserved region.