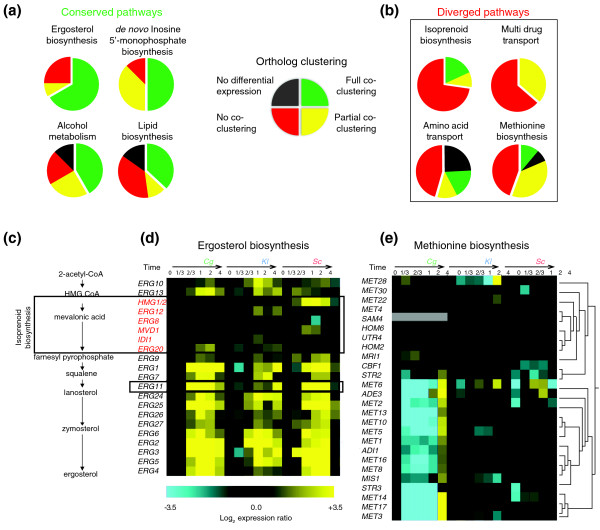
Figure 4.
Pathway expression conservation and divergence. (a) Top conserved and (b) diverged pathway responses as revealed by the soft clustering approach. Each pathway is represented by a pie with four slices - green, yellow, red, and black - denoting the percentage of orthologs in that pathway for which all three species co-clustered, two species co-clustered, no two species co-clustered, and no species' orthologs were differentially expressed, respectively. Pathways were defined using GO biological process annotations. (c) Schematic of ergosterol biosynthesis, the most conserved pathway response. Interestingly, this pathway includes isoprenoid biosynthesis, for which the response was one of the most divergent. (d) mRNA expression responses of ergosterol pathway genes are shown in order of occurrence in the pathway. Expression levels of genes 3 to 8 (boxed, and red) corresponding to isoprenoid biosynthesis are strikingly divergent. The fluconazole target Erg11 is boxed. (e) Hierarchically clustered mRNA expression responses of methionine biosynthesis genes show extensive divergence across species. Grey expression values denote a gene for which the species lacks an ortholog.