Figure 6.
Synapsis from Centromeres in zip3 fpr3 Cells is Zip2-Independent
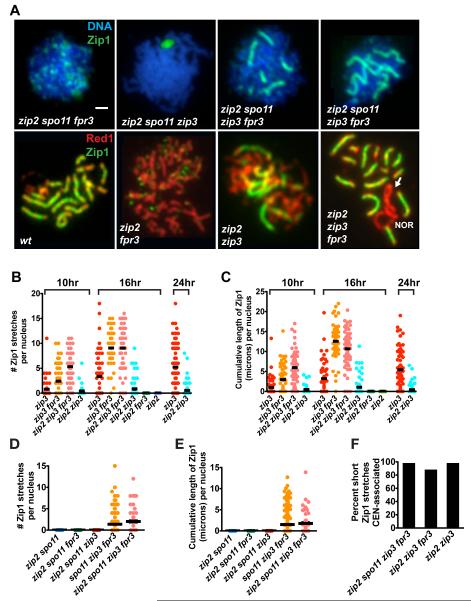
(A) Upper panels show DNA (blue) and Zip1 (green) on spread meiotic nuclei from spo11 mutants. Lower panels show meiotic cores (anti-Red1, red) and anti-Zip1 (green) in spread meiotic nuclei from SPO11+ strains. Arrow points to a zip2 zip3 fpr3 chromosome XII pair with a Zip1-deficient (unsynapsed) region, adjacent to the nucleolar organizing region (NOR). As SC does not span the NOR, synapsis on one side of this chromosomal region would require synapsis initiation at a non-centromeric site. Bar, 1 micron.
(B-E) Scatterplots show the rate and extent of synapsis in zip2 zip3 fpr3 combinations in SPO11+(B and C), or spo11 (D and E) backgrounds. Nuclei were plotted as described in Figure 3G and H; at least 50 nuclei were plotted for each strain. Data shown in (B and C) belongs to a large time course data set, which also includes the data shown in Figure 5; note that wild-type and fpr3 data (shown in Figure 5) are omitted here. At 16 hours, zip2 zip3 fpr3 cells exhibit significantly higher cumulative levels of Zip1 as compared with zip3 cells (two-tailed Mann Whitney p < 0.0001); no significant differences in the cumulative length of Zip1 are apparent between zip2 zip3 fpr3 and either zip3 fpr3 or wild-type strains.
(F) Short Zip1stretches (0.35-0.65 microns in length) exhibited by zip2 zip3 fpr3 nuclei are almost exclusively centromere-associated. 42, 207, and 30 short Zip1 stretches were analyzed for zip2 spo11 zip3 fpr3, zip2 zip3 fpr3, and zip2 zip3 cells, respectively.