Abstract
It is well-established that neurons in the adult mammalian central nervous system (CNS) are terminally differentiated and, if injured, will be unable to regenerate their connections. In contrast to mammals, zebrafish and other teleosts display a robust neuroregenerative response. Following optic nerve crush (ONX), retinal ganglion cells (RGC) regrow their axons to synapse with topographically correct targets in the optic tectum, such that vision is restored in ∼21 days. What accounts for these differences between teleostean and mammalian responses to neural injury is not fully understood. A time course analysis of global gene expression patterns in the zebrafish eye after ONX can help to elucidate cellular and molecular mechanisms that contribute to a successful neuroregeneration. To define different phases of regeneration after ONX, alpha tubulin 1 (tuba1) and growth-associated protein 43 (gap43), markers previously shown to correspond to morphophological events, were measured by real time quantitative PCR (qPCR). Microarray analysis was then performed at defined intervals (6 hours, 1, 4, 12, and 21 days) post-ONX and compared to SHAM. Results show that optic nerve damage induces multiple, phase-related transcriptional programs, with the maximum number of genes changed and highest fold-change occurring at 4 days. Several functional groups affected by optic nerve regeneration, including cell adhesion, apoptosis, cell cycle, energy metabolism, ion channel activity, and calcium signaling, were identified. Utilizing the whole eye allowed us to identify signaling contributions from the vitreous, immune and glial cells as well as the neural cells of the retina. Comparisons between our dataset and transcriptional profiles from other models of regeneration in zebrafish retina, heart and fin revealed a subset of commonly regulated transcripts, indicating shared mechanisms in different regenerating tissues. Knowledge of gene expression patterns in all components of the eye in a model of successful regeneration provides an entry point for functional analyses, and will help in devising hypotheses for testing normal and toxic regulatory factors.
Keywords: neuroregeneration, eye, zebrafish, gene expression analysis, optic nerve, microarrays
Background
It is well known that shortly after development neurons in the mammalian central nervous system (CNS) lose an ability to regenerate following injury.1 A model commonly used to study this phenomenon is the optic nerve because of its accessibility.2 When injured, retinal ganglion cells (RGCs), whose axons comprise the optic nerve, show only a transitory sprouting response and no long distance regeneration to the visual processing areas of the brain.2,3 Nonetheless, some RGCs can extend their axons into a peripheral nerve graft sutured to the cut end of the optic nerve.3 Also, when cultured in a peripheral nerve milieu, RGCs show enhanced regeneration of their axons.4 These observations provide evidence that mature neurons in the mammalian CNS retain some capacity to regenerate their axons under specific conditions. It has been proposed that the extracellular environment of the adult CNS is responsible for the failure to regenerate.5 These detrimental environmental conditions include the infiltration of immune cells from the breakdown of the blood-brain barrier, the release of inflammatory cytokines from immune cells and microglia, inhibitory factors secreted from the myelin, and the formation of a glial scar.6
In marked contrast to mammals, adult teleost fish have the ability to successfully repair axonal injuries in the CNS and regain function. Studies in zebrafish show that transsection of the spinal cord leads to the re-growth of severed axons and the recovery of swimming behavior.7 Likewise, injury to the fish optic nerve results in the re-growth of RGC axons, reestablishment of synapses with topographically correct targets in the optic tectum, and the eventual restoration of vision.8,9
The teleostean optic nerve has been a popular morphological model for the study of regeneration since the 1950's. Following ONX in goldfish, axonal sproutings occur at the cut end of the optic nerve within 3 days, and by 6 days bundles of axonal sproutings penetrate into the injury site.10 The regenerating axons first reach the tectum at 10–12 days after crush and the retinotectal connections are formed between 20 and 40 days post-crush, although the retinotectal topography slowly improves over several months.11,12
Corresponding to morphological events, injury to the fish optic nerve induces a response in the ganglion cells that is associated with the increased biosynthesis of cytoskeletal proteins such as alpha tubulin 1 (tuba1) and growth-associated proteins (GAP) such as growth-associated protein 43 (gap43).13–17 GAPs are transported by the cytoskeletal proteins to the injured end of the axon, where they are incorporated into the membrane of the growth cone.18 As previously measured by semi-quantitative RT-PCR and in situ hybridization in zebrafish, the expression patterns of tuba1 and gap43 define four major phases of the regeneration process. The injury response phase occurs less than 1 day post-ONX during which there is no visible induction of tuba1 or gap43 expression in the RGCs. The preparation for outgrowth phase (1–7 days post-ONX) is marked by the up-regulation of both genes in the RGCs to their maximum levels. The axon extension phase occurs between 5 and 18 days post-ONX during which the up-regulation of tuba1 and gap43 expression begins to decrease. Target contact and synaptic refinement is the last phase of regeneration (14–25 days post-ONX) and is marked by the return of tuba1 and gap43 expression to baseline levels in the RGCs.19–23
To account for differences between mammalian and teleostean responses to neural injury, it has been suggested that the environment surrounding the fish RGCs is permissive, rather than inhibitory, to the growth of axons.7,24,25 How this favorable setting for axonal regeneration is achieved and, in particular, what are the underlying regulatory mechanisms, has yet to be fully understood.
A sequenced genome and the availability of microarrays, morpholino-mediated knockdown technology and many different natural mutants have made the zebrafish an attractive model to study genes and processes involved in damage- induced regeneration in the eye.26–28 To characterize successful optic nerve regeneration, we analyzed the transcriptional response in the zebrafish eye at 6 hours and 1, 4, 12 and 21 days after ONX. These time points span the major phases of regeneration as previously defined by morphological and gene marker criteria. We chose to use the whole eye in order to include possible signaling contributions from the vitreous, glial, immune and vascular elements, as well as the different neural cells of the retina. During the course of our study, Veldman and colleagues29 reported gene expression changes occurring in laser dissected RGCs 3 days after ONX, and Qin et al28 recognized commonalities in the genetic program of light- damaged retinas and surgically damaged zebrafish heart30 and fin.31 Comparison of our data set with results of these earlier studies confirms and extends these analyses.
Methods
Zebrafish and ONX
Wild type adult male zebrafish, Danio rerio, were obtained from a commercial supplier (Ekkwill, Gibsonton, FL) and maintained in 30 gal aquaria at 28 °C on a 14:10 light-dark cycle.32 For optic nerve crush fish were anesthetized by immersion in 0.033% aminobenzoic acid ethylmethylester (MS222; Sigma, St. Louis, MO).14,29 The left optic nerve was exposed by gently pulling the eye out of the orbit and then crushing the nerve with forceps. Care was taken not to injure the blood vessel running along the optic nerve. If bleeding was visible following the surgery the fish was removed from the study. The eye was then gently replaced in the orbit. As a SHAM control the right eye of the same fish was gently pulled out of the orbit and replaced with the nerve remaining intact. The fish were then placed in a recovery tank. At timed intervals post-crush (6 hours and 1, 4, 12, and 21 days for microarray analysis; additional time points for qPCR analysis) the fish were euthanized by overdose of MS-222 and the eyes (including retina, lens, vitreous, anterior and posterior segments) removed, flash-frozen on dry ice and stored at −70 °C. For microarray analysis two biological replicates (2 pools with 5 eyes/pool for each time point) were prepared. Validation with qPCR was performed using three additional independent biological replicates (3 pools with 4–5 eyes/pool at each time point and condition). All animals were treated according to the guidelines of the Internal Animal Care and Use Committee of Boston University.
RNA extraction and reverse transcription (RT)
Pooled zebrafish eyes were homogenized in Tri Reagent (Sigma) and total RNA was extracted as previously described33 and treated with DNase I (Roche, Indianapolis, IN). RNA prepared for microarray analysis was further purified using the RNeasy Mini Kit RNA cleanup protocol (Qiagen, Germantown, MD). An aliquot of each extract was used for spectrophotometry to determine RNA quality and concentration. RNA with a 260/280 ratio between 1.95–2.2 and a 260/230 ratio >1 and <3 was considered satisfactory and was used in this study. Each RNA extract was assayed in triplicate and an average value was determined. A 1 μg aliquot was taken of each sample and electrophoresed on an agarose gel to confirm quality and concentration. cDNA was synthesized from total RNA (3 μg; 20 μl final reaction volume) with oligo(dT) priming using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. A minimum of two RT reactions was performed for each biological replicate for technical replicate comparison.
Microarray preparation and analysis
Independent hybridizations of two biological replicates at each time point and treatment condition (SHAM/ONX) were performed. Processing and hybridization of the RNAs was performed by the Boston University Microarray Core Facility, Boston University MA. Total RNA was converted to biotinlabeled cRNA using Gene Chip One-Cycle Target labeling kit and hybridized to the zebrafish genome array according to the manufacturer's guidelines (Affymetrix, Santa Clara, California, United States). Genes that were considered differentially expressed at at least one time point were identified by a q value of less than 0.15 (6 hrs, 1, 4, 12, 21 days post-ONX versus time-matched SHAM), fold change of greater than 2, and Microarray Suite (MAS) 5.0 absent-present call. The differentially expressed probe sets were annotated based on NCBI's UniGene ID Resource using the Affymetrix NetAffx Analysis Center. Gene Ontology category assignments were determined using NCBI's EntrezGene (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene), Ensembl (www.ensembl.org) and Onto-Express (http://vortex.cs.wayne.edu/ontoexpress). Hierarchical clustering with the average linkage method was performed using Cluster software and results were visualized using TreeView.34 The full gene expression data set described here is accessible through NCBI's Gene Expression Omnibus (GEO) database (accession number: GSE19298).
qPCR and data analysis
qPCR was performed on an ABI Prism 7900HT sequence detection system (Applied Biosystems) with SYBR green fluorescent label. Sample preparation, cycling parameters, and validation of elfa (eukaryotic elongation factor alpha) as a normalizer were performed as previously described.35 For each sample a dissociation step was performed at the end of the amplication phase to identify a single, specific melting temperature for each primer set. PCR was performed twice on each sample for a minimum of 12 data sets generated for each sample/gene combination (2–3 biological replicates × 2 RT reactions × 1–2 PCR runs × 3 reactions per PCR run). Data generated by qPCR were compiled and collected using SDS 2.2 software (Applied Biosystems). Data were exported to QGene to determine the PCR amplification efficiency (E) for each primer pair where E = 10(−1/slope) as determined by linear regression analysis of a dilution series of reactions.36 All amplifications had a PCR efficiency value between 1.83 and 2.3. Cycle threshold (Ct) values were adjusted for differences in efficiency of each primer set/gene target and normalized to elfa as an internal reference (NE = (EREF) CtREF/(ETARGET)CtTARGET). Normalized gene expression data from biological replicates were averaged and shown as fold-change of ONX/SHAM ± standard error of mean (SEM). Statistical analysis was performed using the Sigma-Stat 2.0 package (Aspire Software, Leesburg, VA). Data were analyzed by oneway analysis of variance (ANOVA) followed by the Tukey method for pair-wise multiple comparisons to determine significance with P < 0.05.
Oligonucleotides
All oligonucleotide primers were synthesized by Invitrogen. Gene-specific oligonucleotide primers were developed using Primer Express software (Applied Biosystems) except for elfa primers which were previously published.37 All primer sets spanned an exon-exon junction to avoid errors due to contaminating genomic DNA. Primer sets were tested for specificity using standard RT-PCR and zebrafish embryo cDNA as template to verify production of a single band of the predicted size. Sequences for all primers used in this study as well as gene names, gene abbreviations and NCBI accession numbers can be found in Table 1.
Table 1.
Oligonucleotides primers used for qPCR analysis of selected transcripts from microarrays.
| Gene name | Gene symbol | Sequence (5′-3′) | Accession # |
|---|---|---|---|
| keratin 18 | krt18 | F) GGTCCTGATGTCCACGACTACA R) GGAGCACTAGACGAGCATTGTTT |
NM_178437.2 |
| myeloid specific peroxidase | mpx | F) GGACCACACCCCTCATAAACA R) CCAGCGGGCAAATGGA |
NM_212779 |
| glyceraldehyde-3-phosphate dehydrogenase | gapdh | F) GTGGAGTCTACTGGTGTCTTC R) GTGCAGGAGGCATTGCTTACA |
BC083506 |
| cathepsin C | ctsc | F) CTTCCTCAACACTGGGATTGG R) CCATAGTGGCGAAAGAGTAGCA |
NM_214722 |
| monoamine oxidase | mao | F) TGGCTGCTATACCGCCTACTTC R) CCGTTTCAGTTCCTGCAAAGTAC |
NM_212827.2 |
| synaptoporin | synpr | F) CAGGAACCCATCTGGACCAA R) TATTACCAGCCCAAAGTAAGAAGTTCA |
NM_001004532 |
| elongation factor 1-alpha | elfa | F) CTTCTCAGGCTGACTGTGC R) CCGCTAGCATTACCCTCC |
AY422992 |
| parvalbumin 2 | parvalb2 | F) TGCTGAGACCAAGGCTTTCC R) CCATGAATGCTTAGGCCTTTACA |
NM_131516 |
| parvalbumin 4 | parvalb4 | F) TCTGACGGAGACGGCAAGAT R) AAGTTGTTCCTTCGGAGCAAGA |
NM_212783 |
| fIbronectin 1 | fn1 | F) CTCGTGTCAAAGGAGAAATCACAA R) GCGCCAGGTCAGAGTGATG |
NM_131520 |
| integrin, alpha 5 | itga5 | F) GCTGACTGTGCCGAATGGA R) GAGCAACGAAGATGGGAGACA |
NM_001004288 |
| phosphoglycerate mutase 2 | pgam2 | F) AGGACCATCCATATCACAAGATCA R) GGGCAGCTCACCCTCTTTC |
NM_201024 |
| cyclin e | ccne | F) CGCAGTATGCATCAGAAAGCA R) GAGCAGGTTGTTCCAAACTTCAT |
NM_130995 |
| cholinergic receptor, nicotinic, beta polypeptide 3a | chrnb3a | F) CAGACACATCCGGAAGGAACA R) CCAACACCTGAGCCACAAATT |
NM_201220 |
| glycine receptor, alpha 4b | glra4b | F) GCAGGAGCAACGCAACAGA R) CTCGGTGTGCCTCCTGTGA |
XM_684976 |
| granulin 1 | grn1 | F) TACTGCGATGCTCAAACTGT R) CTGCAACACTGACCCATTGG |
NM_001020802 |
| cryptochrome 3 | cry3 | F) TTTAGGACTCAGGGTGACAGCTT R) TTGCTACCCAGGCCTTCCTAT |
NM_131786 |
| kinetochore associated 2-like | kntc2l | F) ACGAGGAGGTCAACCTGTCTAAAA R) GCTGATAGCGGGATGAGCTT |
NM_001003863 |
| growth associated protein 43 | gap43 | F) CAGCCGACGTGCCTGAA R) TCCTCAGCAGCGTCTGGTTT |
NM_131341 |
| alpha tubulin 1 | tuba1 | F) GGAGCTCATTGACCTTGTTTTAGATA R) GCTGTGGAAGACCAGGAAACC |
NM_194388 |
| caspase 8, apoptosis-related cysteine peptidase | casp8 | F) GATCGAGAGGTTCAGGAATCAGA R) CATTGTTTCAGATACAGGGTTGTTG |
NM_131510.2 |
| BCL2-antagonist of cell death | bad | F) CGGCCAACAGCTGAGAAGA R) GCTGGGCGATTGACTCATCT |
NM_131579.1 |
Results
Expression of axogenesis markers after optic nerve crush
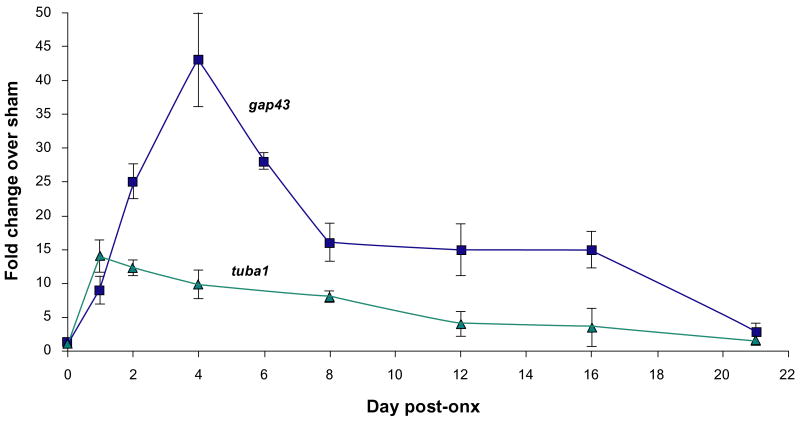
To verify the time course of optic nerve regeneration in our fish, the expression of the known markers of axogenesis, gap43 and tuba1, was measured at defined intervals after ONX over a 3-week period (Fig. 1). Both gap43 and tuba1 showed no induction at the earliest time point (6 hr) but were up-regulated significantly by 1 day following ONX. Whereas the up-regulation of tuba1 peaked at 1 day after ONX, gap43 reached a maximum at 4 days. Both markers remained elevated over SHAM controls throughout the remaining 21 days. These expression patterns strongly correlate to previous studies measuring axon outgrowth and tuba1 and gap43 mRNA levels.20,23,38 Using these expression patterns as a guide, time points for microarray analysis were selected as follows: 6 hours post-ONX (injury response phase), 1 day post-ONX (injury response/preparation for outgrowth phases), 4 days post-ONX (preparation for outgrowth phase), 12 days post-ONX (axon extension phase) and 21 days post-ONX (target contact/synaptic refinement phase).
Figure 1. Expression of established gene markers of optic nerve regeneration as a function of time after ONX, as determined by qPCR analysis.
Each data point represents the mean fold change (ONX/SHAM) +/− SEM of 3 independent biological replicates. Both genes showed significant differences across regeneration stages by one-way ANOVA P < 0.05.
Global gene expression analysis following optic nerve crush
Microarray analysis was performed with biological duplicates at each time point. The gene expression data were represented as fold change of ONX over SHAM samples. Table 2 lists the probe sets that were altered at least 2-fold at each time point and the full list of transcripts altered throughout the time course can be found in Table S1 (supplementary data). Of a total of 14,900 probes on the array, 309 were significantly changed compared to SHAM as early as 6 hr post-ONX, but the number of up- or down-regulated transcripts at 4 days was 7- to 10-times greater than at all of the other time points. To better visualize the number and magnitude of the expression changes as a function of time after ONX, the transcripts altered at each time point were plotted against their corresponding fold changes (Fig. 2). This plot shows that for four of the time points (6 hours, 1, 12, and 21 days) the majority of the induced transcripts were up-regulated between 2- and 10-fold. By comparison, during the preparation for outgrowth at 4 days there were not only a greater number of genes up-regulated, but also a large percentage (>20%) of the transcripts were altered more than 10-fold. By contrast the number and magnitude of down-regulation of transcripts were similar at all time points with the majority of transcripts reduced 2- to 10-fold. A number of the most up- and down-regulated transcripts were incompletely annotated or were novel sequences; therefore, a list of the top ten most altered probe sets that corresponded to known genes was compiled (Tables 3 and 4). Several genes were among the most altered genes at more than one time point. Of the ten most up-regulated genes at 6 hours, 6 were also found at 24 hours after ONX, demonstrating that the pattern of transcription during the injury response phase substantially overlapped into the preparation for outgrowth phase. By contrast, comparison of the top ten most up-regulated genes at 4, 12 and 21 days revealed distinct phase-related expression profiles.
Table 2.
Transcripts up- or down-regulated at least 2-fold at defined intervals after ONX as compared to SHAM.
| 6 hours | 1 day | 4 days | 12 days | 21 days | |
|---|---|---|---|---|---|
| up-regulated transcripts | 185 | 283 | 2102 | 342 | 230 |
| down-regulated transcripts | 124 | 142 | 1302 | 209 | 291 |
Figure 2. Transcript abundance at each time point following ONX.
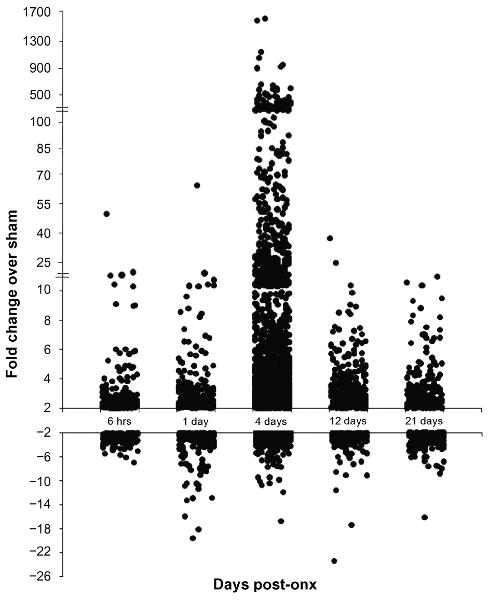
Plot shows individual probe sets from each time point up- or down-regulated at least 2-fold. Note the breaks in the scale on the positive y-axis.
Table 3.
Top ten up-regulated genes identified at each time point after ONX compared to SHAM.
| Probe ID | Gene name | Fold change | |
|---|---|---|---|
| 6 hours | Dr.19525.2.A1 | Quinoid dihydropteridine reductase a | 16.1 |
| Dr.17738.2.A1 | tumor necrosis factor, alpha-induced protein 6 | 15.9 | |
| Dr.15281.1.A1 | tissue inhibitor of metalloproteinase 2, like | 11.3 | |
| Dr.10314.1.S1 | matrix metalloproteinase 13 | 9.1 | |
| Dr.12986.1.A1 | v-fos FBJ murine osteosarcoma viral oncogene homolog | 5.9 | |
| Dr.20198.1.S1 | heat shock cognate 70-kd protein | 5.8 | |
| Dr.14282.1.S1 | activating transcription factor 3 | 4.7 | |
| Dr.967.1.S1 | matrix metalloproteinase 9 | 4.1 | |
| Dr Affx.2.25.A1 | granulin 1 | 4.0 | |
| Dr.314.1.S1 | achaete-scute complex-like 1a | 4.0 | |
| 1 day | Dr.17738.2.A1 | tumor necrosis factor, alpha-induced protein 6 | 63.2 |
| Dr.17618.1.A1 | kininogen 1 | 10.9 | |
| Dr.15281.1.A1 | tissue inhibitor of metalloproteinase 2, like | 10.2 | |
| Dr.25168.1.S1 | asparagine synthetase | 10.0 | |
| Dr.14282.1.S1 | activating transcription factor 3 | 8.6 | |
| Dr Affx.2.25.A1 | granulin 1 | 8.5 | |
| Dr.5094.3.A1 | Smu-1 suppressor of mec-8 and unc-52 homolog | 8.3 | |
| Dr.92.1.A1 | growth associated protein 43 | 7.4 | |
| Dr.12986.2.S1 | v-fos FBJ murine osteosarcoma viral oncogene homolog | 5.9 | |
| Dr.19525.2.A1 | Quinoid dihydropteridine reductase a | 5.7 | |
| 4 days | Dr.4412.3.A1 | spectrin alpha 2 | 1638.3 |
| Dr.19902.2.A1 | cathepsin | 1088.0 | |
| Dr.2596.2.A1 | MpV17 transgene, murine homolog, glomerulosclerosis | 999.7 | |
| Dr.12259.1.S1 | B-cell translocation gene 4 | 849.7 | |
| Dr.5719.1.A1 | claudin d | 828.0 | |
| Dr.728.1.S1 | pseudouridylate synthase 1 | 560.9 | |
| Dr.4412.2.S1 | dentin sialophosphoprotein preproprotein | 477.4 | |
| Dr.20877.1.S1 | importin alpha | 464.6 | |
| Dr.4039.3.S1 | crystallin, zeta (quinone reductase) | 464.3 | |
| Dr.24219.4.S1 | cellular nucleic acid-binding protein | 410.0 | |
| 12 days | Dr.268.1.S1 | ependymin | 35.3 |
| Dr.4412.3.A1 | spectrin alpha 2 | 9.8 | |
| Dr.7753.1.S1 | parvalbumin 5 | 8.9 | |
| Dr.22670.1.A1 | calpain 8 | 7.3 | |
| Dr.12665.1.S1 | G protein-coupled receptor 172-like | 6.6 | |
| Dr.13204.1.A1 | transcription factor IIIA-like | 6.4 | |
| Dr.26388.1.S1 | mitotin | 6.3 | |
| Dr Affx.1.17.S1 | nuclear receptor subfamily 5, group A, member 1b | 6.2 | |
| Dr.588.1.S1 | forkhead box A sequence | 5.7 | |
| Dr.20821.1.A1 | plasminogen | 5.5 | |
| 21 days | Dr.14499.1.S1 | globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like | 15.3 |
| Dr.23423.2.S1 | zona pellucida glycoprotein 2.2, 2.3 | 8.8 | |
| Dr.3073.1.A1 | serpin peptidase inhibitor, clade A, member 7 | 7.9 | |
| Dr.14434.1.S1 | piwi-like 1 | 7.0 | |
| Dr.20821.1.A1 | plasminogen | 5.5 | |
| Dr.6031.2.A1 | polymerase (DNA directed), lambda | 4.7 | |
| Dr.8107.1.S1 | SRY-box containing gene 17 | 4.5 | |
| Dr.8280.1.S2 | decapentaplegic and Vg-related 1, RNA binding protein | 4.4 | |
| Dr.2452.2.A1 | complement component 9 | 4.3 | |
| Dr.19877.1.S1 | V-set and transmembrane domain containing 2 like | 4.2 | |
Table 4.
Top 10 down-regulated genes identified at each time point after ONX as compared to SHAM.
| Probe ID | Gene name | Fold change | |
|---|---|---|---|
| 6 hours | Dr Affx.1.80.S1 | parvalbumin 4 | −7.0 |
| Dr.26411.2.S1 | troponin I type 2 | −5.5 | |
| Dr.20990.2.S1 | titin | −5.0 | |
| Dr.8472.1.S1 | troponin C | −4.8 | |
| Dr.13621.1.A1 | phosphofructokinase | −4.6 | |
| Dr.13621.1.A1 | ATPase, ca++ transporting, 1 | −4.3 | |
| Dr.11552.1.S1 | ATPase, ca++ transporting, 1 like | −4.2 | |
| Dr.4812.1.S1 | myosin, heavy polypeptide 1 | −4.1 | |
| Dr.24260.1.S1 | myosin, light polypeptide 3 | −4.0 | |
| Dr.10620.1.S1 | troponin T3b | −3.8 | |
| 1 day | Dr.26517.1.S1 | phosphoglycerate mutase 2 | −9.0 |
| Dr.18267.1.S1 | creatine kinase | −8.4 | |
| Dr.10620.1.S1 | troponin T3b | −8.3 | |
| Dr.24260.1.S1 | myosin, light polypeptide 3 | −8.0 | |
| Dr.24941.1.S1 | nucleoside diphosphate kinase-Z2 | −7.2 | |
| Dr.4200.1.A1 | slow myosin heavy chain 1 | −7.1 | |
| Dr.13621.1.A1 | phosphofructokinase | −6.5 | |
| Dr.2914.1.S2 | myosin, light polypeptide 2 | −6.1 | |
| Dr.6818.1.S1 | myelin protein zero | −5.9 | |
| Dr.460.1.A1 | parvalbumin 2 | −5.7 | |
| 4 days | Dr.18462.1.A1 | iroquois homeobox protein 4b | −9.9 |
| Dr.26132.1.S1 | phosphoenolpyruvate carboxykinase 1 | −9.5 | |
| Dr.4907.1.S1 | fibrinogen, gamma polypeptide | −6.6 | |
| Dr.861.1.S1 | dopa decarboxylase | −6.6 | |
| Dr.4520.1.A1 | N-myc downstream regulated gene 4 | −5.7 | |
| Dr.14053.1.A1 | synaptoporin | −5.5 | |
| Dr.7232.1.S1 | homeo box B8a | −5.3 | |
| Dr.25364.1.A1 | hexokinase 1 | −5.2 | |
| Dr.11068.1.A1 | calbindin 2 | −4.9 | |
| Dr.15720.1.S1 | connexin 55.5 | −4.8 | |
| 12 days | Dr.4865.1.A1 | serine (or cysteine) proteinase inhibitor, clade D, member 1 | −23.3 |
| Dr.12235.1.A1 | Pyrroline-5-carboxylate reductase-like | −9.1 | |
| Dr.15445.1.S1 | small nuclear RNA activating complex 1 | −6.2 | |
| Dr.24309.1.S1 | troponin T1 | −6.0 | |
| Dr.5733.1.S1 | homeo box c8a | −5.4 | |
| Dr.14434.1.S1 | piwi-like 1 | −4.8 | |
| Dr.6386.1.A1 | suppressor of variegation 3–9 homolog 1 | −4.5 | |
| Dr.2596.2.A1 | glomerulosclerosis | −4.3 | |
| Dr.16916.1.A1 | Ankyrin 1 | −4.2 | |
| Dr.26132.1.S1 | phosphoenolpyruvate carboxykinase 1 | −4.1 | |
| 21 days | Dr.3201.1.S1 | zinc finger-like gene 2a | −6.8 |
| Dr Affx.2.5.S1 | dead end | −6.8 | |
| Dr.2059.1.A1 | solute carrier family 2 (facilitated glucose transporter), 2 | −6.1 | |
| Dr.21414.1.A1 | homeo box B9a | −6.0 | |
| Dr.26360.1.A1 | glycoprotein hormones, alpha | −5.3 | |
| Dr.14034.1.A1 | NOD3 protein | −5.1 | |
| Dr.17906.1.A1 | Zinc finger, BED domain containing 4 | −4.7 | |
| Dr.179.1.S2 | T-box gene 16 | −4.6 | |
| Dr.591.1.S1 | forkhead box A1 | −4.6 | |
| Dr.25698.1.S1 | LIM homeobox 8 | −4.4 | |
Validation of microarray analysis withq PCR
To validate the microarray expression data, qPCR was performed on a subset of genes representative of two different temporal patterns (Fig. 3). Four genes, monoamine oxidase (mao), synaptoporin (synpr), cryptochrome 3 (cry3) and one cut like 1 (onecutl) were down-regulated transiently between 6 hours and 4 days post-ONX and their expression returned to SHAM control levels or increased slightly thereafter (Fig. 3a). The second set of genes, cathepsin C (ctsc), myeloid peroxidase (mpx), granulin 1 (grn1), and keratin 18 (krt18) were markedly up-regulated in the injury response/preparation for outgrowth phase of regeneration and subsequently returned to baseline levels (Fig. 3b). As shown in the right-hand panels, the qPCR expression of all eight genes in biological replicates strongly resembled the pattern found on the microarray (Fig. 3b). The expression patterns of twentythree additional genes selected from the microarray were also measured by qPCR (Table S2, supplementary data). These genes were selected on the basis of their direction or magnitude of change, their potential functional importance, or because they are genes of interest in our laboratory.35 An additional twelve genes selected for validation were representative of six different gene ontology categories (Figs. 4 and 5; see below). Including gap43 and tuba1, more than 90% (41/45) of tested transcripts identified as up- or down-regulated on the microarray were successfully validated by qPCR using biological replicates.
Figure 3. Comparison of gene expression profiles by microarray and qPCR analysis.
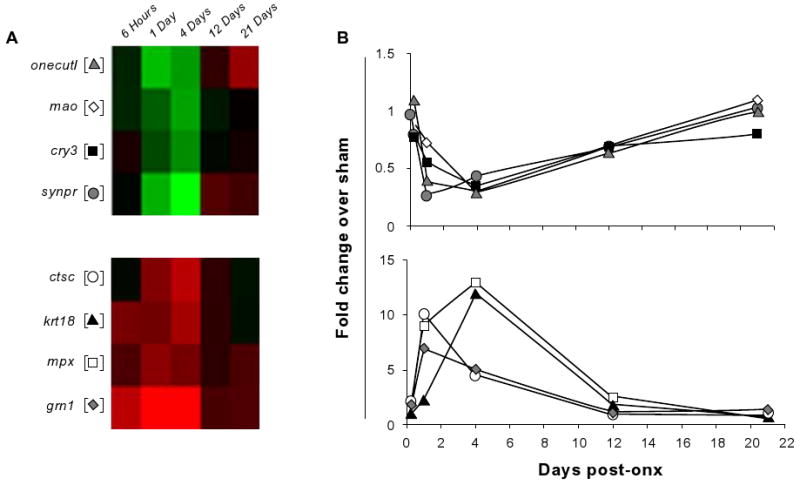
The time course of expression changes of selected genes as determined by the microarray analysis (A) and qPCR (B). in (A) two clusters represent genes down-regulated (top panel) or up-regulated (lower panel) during the early phase of regeneration. (B) The qPCR expression profiles of the selected target genes are very similar to the microarray-based profiles. Microarray results are plotted using the standard red/green (up/down) coding and mean (n = 3) fold change of SHAM/ONX is plotted for the qPCR analysis.
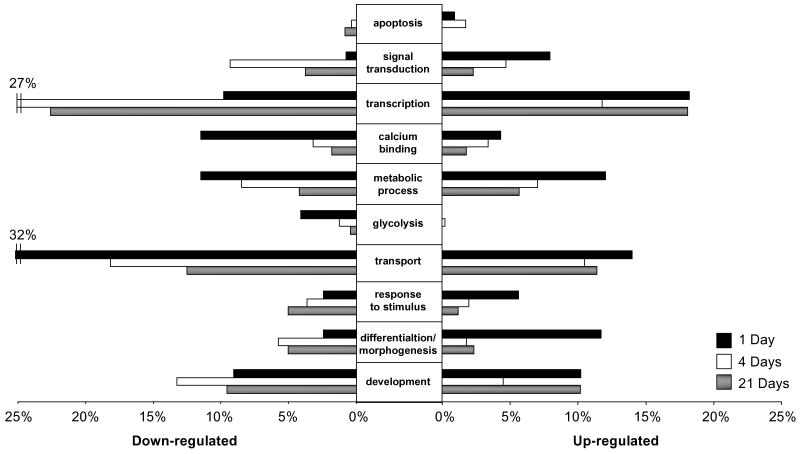
Figure 4. Gene ontological analysis of changes in gene expression after ONX.
Selected gene ontology biological process or molecular function categories. Percentages represent the number of genes regulated in one gene ontology group compared to all genes differentially regulated at that time point (1, 4, or 21 days). Ontological groups were chosen according to whether each gene class was statistically over-represented using Onto-Express (with a hypergeometric distribution and an FDR correction (P < 0.05)) in up-regulated or down-regulated genes in at least one time point.
Figure 5. Cluster analysis and target gene expression.
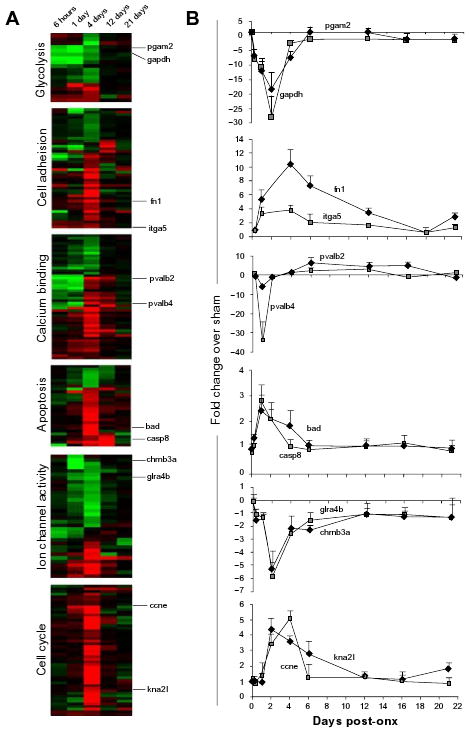
Cluster analysis was performed on microarray data using cluster and heat maps were constructed using TreeView. Two genes from each gene ontology class were selected for qPCR analysis at nine time-points (0 hrs, 6 hrs and 1, 2, 4, 6, 12, 16 and 21 days). The results are represented as mean fold-change of ONX over SHAM. Error bars show standard error of mean (S.E.M) of biological triplicates. For expanded heat maps, see Fig. S1.
Gene ontology analysis of gene expression changes during optic nerve regeneration
The differentially expressed genes were categorized by gene ontology (GO). This analysis resulted in a large number of overlapping gene groups that were defined by various biological processes and molecular functions (Fig. 4). These broad functional categories contained hundreds of identifiable genes; therefore, this analysis organized the data set into more specific functional groupings, in order to provide a broad depiction of the salient processes involved in optic nerve regeneration. As the 6 hour GO analysis strongly resembled the 1 day analysis and the 12 day time-point resembled either the 4 or 21 day analysis, for clarity only the 1, 4, and 21 day time points are shown. Broad categories like transcription and transport made up a large percentage of the total up-and down-regulated transcripts, with 27% of the transcripts down-regulated at 4 days involved in transcription and 32% of the down-regulated transcripts at 1 day involved in transport. Several categories also showed temporally regulated expression patterns. As regeneration progressed, the number of transcripts involved in stimulus response and cell differentiation/tissue morphogenesis showed decreasing up-regulation and increasing down-regulation. The number of transcripts associated with calcium binding and glycolysis showed a strong down-regulation at 1 day that diminished at later time points. Transcripts involved in apoptosis comprised only a small proportion of the total regulated transcripts but there was a clear initial induction of transcripts involved in the process followed by down-regulation by the end of regeneration.
Hierarchical clustering of functional groups identifies up- and down-regulated genes during regeneration
Several gene ontology functional groups were selected for further analysis by hierarchical clustering of the differentially expressed genes in each group and distinct temporal expression patterns emerged (Fig. 5a). Heat maps from six of these GO classes are presented in Figure 5. In order to validate these expression patterns and develop a more specific time-frame for the regulation of these processes, qPCR was performed on two target genes from each cluster at nine time points: 0 hours, 6 hours, 1, 2, 4, 6, 12, 16, and 21 days post-ONX (Fig. 5b). Many genes involved in glycolysis were down-regulated during the injury response and preparation for outgrowth phases of regeneration, with little change in expression levels during axon extension and synaptic refinement (Fig. 5a). Genes gapdh and pgam2 which encode glycolytic enzymes, were steeply down-regulated in response to injury and their expression increased at later time points with gapdh leveling off before pgam2 (Fig. 5b). The expression of a number of genes involved in cell adhesion changed little until the preparation for outgrowth phase began (4 days post-ONX; Fig. 5a). Exceptions were the cell adhesion gene fibronectin 1 (fn1) and its receptor integrin (itga5). Both were up-regulated from the injury response through axon extension phases, between 6 hours and 12 days (with fn1 > itga5), and their expression then decreased and stabilized during the phase when target contacts are known to be established (Fig. 5b). A large cluster of genes encoding calcium binding proteins were down-regulated in response to injury (6 hours and 1 day post-ONX) but increased in expression during outgrowth preparation (4 days) before leveling off during phases defined by axon extension and synaptic refinement. Two members of the parvalbumin family of calcium binding proteins were assayed by qPCR (Fig. 5b). The expression of pvalb2 and pvalb4 was strongly repressed at 1 day (∼5-fold and 35-fold, respectively). Their expression then increased between 4 and 12 days (pvalb2 remained up-regulated through 16 days) and returned to approximately baseline levels by 21 days. The majority of genes involved in apoptosis were unchanged at all phases of regeneration except during preparation for outgrowth (4 days), where more than 80% of genes in this category were up-regulated (Fig. 5a). The expression of pro-apoptotic genes caspase 8 (casp8) and BCL-2 antagonist of cell death (bad) showed a sharp increase between 1 and 4 days and transcript levels returned to baseline thereafter (Fig. 5b). Many genes with ion channel activity were down-regulated during injury response and preparation for outgrowth phases (between 6 hours and 4 days), but a smaller set were induced in this early phase of regeneration (Fig. 5b). The expression of cholinergic and glycine receptors (chrnb3a and glra4b) was down-regulated very briefly between 2 and 4 days and then progressively returned to SHAM control levels as regeneration progressed (Fig. 5b). The expression of genes encoding components of the cell cycle was mainly unchanged at all stages of regeneration except the phase of outgrowth preparation (4 days), where almost all transcripts in this category were up-regulated (Fig. 5a). The genes cyclin E (ccne) and kinetochore-associated 2 like (kna2l) were representative of the restricted temporal expression patterns of genes in this category, with an up-regulation between 2 and 4 days; however, kna2l remained induced through 6 days (Fig. 5b). Expanded heat map clusters with full gene annotations can be found in Figure S1 (supplementary data).
Comparison with other regeneration transcriptional profiles
In order to help elucidate the possible cellular origin of the observed transcriptional changes, we compared our dataset with one from another study which examined the gene expression changes in laser-dissected RGCs at 3 days post-ONX.29,30 Using our 4 day time-point for comparison, we found that 86% of the 313 transcripts up-regulated in the RGCs 3 days after ONX were also induced in the whole eye at 4 days (Table S3, supplementary data). Of the top 20 most up-regulated transcripts in isolated RGCs, 19 were induced more than 2-fold in the whole eye. In addition, all 29 transcripts down-regulated in the RGCs were also reduced in the whole eye. Although technical differences in the microarray analyses in the two studies could account for some difference in fold up- or down-regulation, it is worth noting that plasticin, a gene known to be expressed in RGC,39 was induced 30-fold in isolated RGC but only about 3-fold in eye. By contrast, induction of beta thymosin in RGC was about 60% of that in eye (29- vs. 49-fold), suggesting an additional cellular source. Also, whereas the measured induction of SRY-box containing gene 11b (sox11b) was similar in RGC and eye (9 and 10-fold, respectively), the SRY-box containing gene 11a (sox11a) transcript was induced to higher levels in RGC than eye (8 and 2.5-fold, respectively), implying that the two sox11 paralogs in zebrafish have evolved different expression domains, regulation or function. Another study recently published by Saula et al looked at expression differences in the retina 24 hours after optic nerve injury or sham-treatment identified an up-regulation of atf3 in the retinal ganglion cells, the nerve fiber layer and the optic nerve of the injured eye.40 Our study also recognized a regulation of atf3 in the injured eye, with initial up-regulation at 6 hours post-ONX (4.7-fold), further induction at 1 day post-ONX (8.6-fold), and maximal up-regulation at 4 days post-ONX (16.8-fold; Table S1).
To determine whether optic nerve regeneration shares common mechanisms with zebrafish heart and fin regeneration, we compared the gene expression profiles in the present study with those reported for heart and fin regeneration.30,31 Of the 829 and 662 transcripts differentially expressed in fin and heart regeneration, respectively, there were 132 overlapping transcripts, 119 up-regulated and 13 down-regulated. When these transcripts were compared to our regenerating optic nerve expression profile we found 68 up-regulated and 6 down-regulated transcripts commonly expressed among all regeneration models (Table S4, supplementary data). Gene ontology analysis of these transcripts revealed that nearly a third were involved in cell cycle or cell adhesion functions, with remaining transcripts implicated in inflammation (3%), metabolism (9%), protein folding (7%), transport (7%), structural components (4%), and other/unknown processes (38%). A number of these commonly regulated genes were among the most induced genes in our dataset including granulin 1, activating transcription factor 3, embryonic transglutaminase, and TNF alpha-induced protein 6 (Table 3).
Discussion
This study demonstrates that optic nerve damage induces multiple, phase-related transcriptional programs in zebrafish eye over the 21 day regeneration period. In general, the global gene expression patterns resemble what is seen with the established neuroregeneration markers gap43 and tuba1, with the maximum number of genes changed and the highest fold-change occurring between 1 and 4 days and smaller and more selective changes at later time points. It would be interesting to determine the corresponding time course pattern of genes expressed in the tectum where functional input is initially lost and by 21 days reformed and remodeled.22,41 On microarrays the two established markers show the expected pattern and, using qPCR, we are able to validate more than 90% (41/45) of selected transcripts identified as up- or down-regulated (see Results). This high success rate is good evidence that the data from the microarrays accurately represent the expression patterns of the different functional groups.
Regulation of cellular processes during regeneration
Transcripts that were altered belong to many different cell types and gene ontology classes, indicating a broad impact of optic nerve injury on cellular functions of the eye. Further analysis of some of these functional groups reveals some unexpected cellular mechanisms altered by neural injury.
Apoptosis and cell proliferation
Apoptosis and cell-cycle-related gene regulation demonstrates an interesting pattern which shows early periods of cell death-associated gene up-regulation following injury and a subsequent increase in cell proliferation-associated gene expression (Figs. 4 and 5). Although this phenomenon remains mostly unexplored in zebrafish during regeneration, there have been reported changes in the number and distribution of RGCs after ONX injury.42 In goldfish, studies show that 10%–20% of the RGCs are lost during a wave of cell death after ONX, which is a small number compared to mammals where nearly all RGCs undergo atrophy/apoptosis after injury.43–45 Another study using in vitro organotypic culture of adult zebrafish retina reported a rate of nearly 50% RGC apoptosis as determined by TUNEL staining of the retinal explants after 10 days in culture.46 This apoptosis has been attributed to the axotomy required to prepare the retinal explants.47 The loss of RGCs in the explants do not appear to be compensated by cell proliferation as determined by BrdU uptake and labeling, although extensive proliferation is observed among other cell types.46 These differences between in vivo and in vitro findings suggest that signals that are present in the intact organism, and lacking in the explanted retina, may be responsible for RGC survival and activation of axonal re-growth after ONX. Nonetheless, our gene expression findings suggest that there may indeed be a low rate of apoptosis following injury as well as an increase in cell proliferation. Whether proliferation is required to replace RGCs lost to apoptosis, or is necessitated by another cell type or another process such as DNA repair rather than proliferation per se will require further study.
Energy metabolism
One of the most striking results from the expression dataset is the near complete shutdown of glycolysis-related gene expression during the early stages of optic nerve regeneration (Figs. 4 and 5). It is well-established that neural injury leads to a disruption in blood flow which results in a depleted energy supply to the injured site.48–50 In mammals, one way in which neurons attempt to compensate for this loss is by increasing local glycolytic activity.51 While hyperglycemia can be beneficial to neurons it can also increase astrocytic cell death due to enhanced lactic acidosis.51,52 This disrupted brain glucose metabolism is a characteristic of traumatic brain injury that often predicts a poor prognosis.53 In a study of patients with a subarachnoid hemorrhage, 50% of those who died showed hyperglycosis while all the patients who survived did not.54 The reversal of this process in zebrafish, of reducing the expression of glycolytic enzymes rather than increasing their activity, may be advantageous by preventing the cell death that can result from hyperglycosis.
Calcium signaling and excitotoxicity
The steep down regulation of many calcium binding proteins almost immediately following optic nerve injury is another intriguing result of our analysis (Figs. 4 and 5). A major contributory factor to neuronal damage and death after a neural injury is increased intracellular calcium levels.55 Calcium signals in the form of intracellular calcium increases are sensed and transmitted by Ca+2 binding proteins in the cell.56 Increased calcium levels can lead to pathologic activation of calcium binding proteins, which has been shown to inhibit essential neuronal survival mechanisms in neurodegenerative disease states.57 One interpretation of the reduced expression of calcium binding proteins in zebrafish eye after ONX is that it may diminish the overall impact of injury- induced intracellular calcium levels by impeding calcium signaling pathways. Similarly, the reduction of glutamate and cholinergic receptor expression could be protective, as these receptors are known to be key mediators of excitotoxic cell death following calcium influx.58
Role of other cell types in the RGC environment
Our data show that many transcripts previously shown to be enriched in the lens and anterior segment of the eye (selenoproteins, annexins, collagens)59 are up-regulated following optic nerve crush, while some transcripts highly enriched in the retina (phosducins, glutamate decarboxylases, n-myc, drgl)59 are down-regulated during optic nerve regeneration. This is consistent with the view that gene regulation in cell types other than RGC somehow participate in creating a microenvironment that supports the successful regeneration of the optic nerve. Indeed there is ample evidence in the literature to support this point.
The presence and activity of photoreceptors is known to influence the propensity of ganglion cells to regenerate their axons in rat retinal explants and in the rat model in vivo.60 Secreted signals from photoreceptors may also contribute to optic nerve regeneration. The retinol binding protein purpurin, secreted by photoreceptors, induces neurite outgrowth in retinal ganglion cells in goldfish.61 On the other hand, we observed the down-regulation of numerous opsins involved in phototransduction, an indication that failure of photic transmission due to ONX, or signals emanating from damaged RGC, feedback on normal gene expression in photoreceptors.
Input from amacrine cells is also likely to be important in a successful regenerative response. Amacrine cells in the neonatal rat signal retinal ganglion cells to undergo profound and irreversible loss of intrinsic axon growth ability.62 While the factors involved in this signaling have not yet been elucidated we did identify one family of genes known to be highly enriched in amacrine cells, the calcium binding parvalbumins, as briefly but strongly down-regulated in response to injury in the optic nerve. The function of parvalbumin in neurons is not well understood although decreased expression of parvalbumin is observed in neuronal populations lost early in amyotrophic lateral sclerosis and in GABAergic interneurons in schizophrenia.63,64
Our study reinforces the view that immune cells play a role in optic nerve regeneneration by revealing that numerous macrophage derived factors including chemokines and cytokines (e.g. chemokine C-X-C motif receptors and eosinophil chemotactic cytokine) are induced following ONX. Also, a member of the crystallin family of proteins (i.e. crystallin, zeta), recently shown to stimulate inflammation and regeneration in the eye,65 is one of the most up-regulated genes at our 4 day time point. Immune challenges or injury to the lens and vitreous elicit improved regeneration in mammalian models of optic nerve crush.66,67 Macrophages in particular are thought to secrete factors that promote neuronal survival and axon outgrowth.68
Perhaps the most well-studied factors that facilitate or inhibit optic nerve regeneration are those derived from myelin, reactive glia or fibroblasts.2,6,47,69 Ephrins expressed on astrocyte/fibroblast membranes, semaphorins produced by fibroblasts, netrins expressed on oligodendrocytes, and myelin-derived proteoglycans and neurocans are all known to inhibit adult axon regeneration.69,70 Several genes encoding each of these factors are down-regulated during the injury response and preparation for outgrowth phases of zebrafish optic nerve regeneration (Table S1) suggesting a zebrafish-specific mechanism to prevent this inhibition. There are also beneficial agents expressed in or secreted by glia such as tissue inhibitors or metalloproteinases, fibroblast growth factors, glutamate transporters, and connexins.71–73 Many genes encoding these factors are up-regulated following ONX in the zebrafish (Table S1) signifying additional signaling pathways that support regeneration.
Despite the recognized contribution of diverse cell types in the promotion of neurite outgrowth, many studies focus only on the RGC per se. In particular, cultured RGC are a favored model. A great deal of work has studied RGCs with or without various additives to encourage neurite outgrowth or to neutralize inhibitory signals.74–78 Another study which also utilized the zebrafish ONX model profiled gene expression patterns in isolated RGCs at 3 days post-ONX.29 Results of this study provide a valuable comparison with our dataset (Table S3) as together they can help differentiate the transcriptional response of RGCs from that of other cell types in the eye during regeneration. Since there is ample evidence that mammalian RGCs maintain the intrinsic capacity to regenerate there are likely contributions from other cell types in the eye, including other retinal neurons, oligodendrocytes/astrocytes, and immune cells, which help determine a successful regeneration. A few groups have also investigated genome-wide responses to other types of retinal injury in zebrafish, specifically retinal excision27 and light-induced photoreceptor ablation.28,79 These studies also identified time-dependent changes in global gene expression patterns following injury. Both the light-ablation and retinal excision models involve extensive proliferation of glial cells and neuronal differentiation to replace retinal cells destroyed/excised during the injury, which differs from the injury response to optic nerve crush. However, given the potential role for many cell types during optic nerve regeneration these studies provide a valuable complement to our time course study of optic nerve regeneration.
Common factors in different models of regeneration
Another approach to identify factors that facilitate regeneration is to compare the transcriptional responses in different tissue types that also regenerate. Those factors that are common to all regeneration pathways are likely to play an important role in recovery from injury and in promoting regeneration. Zebrafish are able to regenerate not only their optic nerve but also their heart and caudal fin.30,31 Following partial amputation in both models, the initial response to injury is the formation of a clot or cap around the wound area (between 12–24 hours in fin and 2–3 days in heart).80,81 Regenerative outgrowth occurs when either cardiomyocytes surrounding the wound re-enter the cell cycle (in heart) or a group of progenitor-like cells, believed to derive from de-differentiated mesenchymal cells (in caudal fin) proliferate and migrate to replace the amputated tissue.31,82 The structure and function of the fin and heart are restored around 1–2 weeks and 2 months post-amputation, respectively.30,31,80,81
Of the transcripts identified as expressed in both heart and fin regeneration, more than half are also regulated during optic nerve regeneration (Table S4). A large proportion of the up-regulated transcripts are involved in cell adhesion and cell cycle which indicates the importance of general cellular processes in regeneration. However, in all three models there is very little overlap in the down-regulated transcripts suggesting that signals requiring repression during regeneration may vary in different tissues.
During the course of our study, a report by Qin et al compared gene expression changes in heart, fin and photoreceptor regeneration in zebrafish.28 They identified two genes, hspd1 and mps1, required for fin, heart and photoreceptor regeneration. These two genes are also induced in our optic nerve regeneration model. Collectively, these data indicate that a set of core molecules are regulated during regeneration of zebrafish heart, fin, photoreceptors, and optic nerve and suggest that it may also be productive to search for tissue-specific factors in the non-overlapping expressed transcripts.
Based on commonalities in different regeneration models and paradigms, at least at the molecular level, cross-comparisons are a valid approach to identify important regulatory pathways. For example, treatment with the arylhydrocarbon receptor (AhR) ligand 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin (TCDD) has been shown to impair caudal fin regeneration in zebrafish if exposure occurs within 4 days of amputation.83,84 In mice, TCDD exposure inhibits liver regeneration when administered before, or up to 12 hours after, partial hepatectomy.85 The inhibition of tissue regeneration by TCDD across species suggests that AhR signaling, perhaps activated by endogenous ligand, interacts negatively with critical regenerative pathways. A recent study using morpholino technology established that the inhibition of zebrafish caudal fin regeneration by TCDD is dependent on the AhR and its heterodimeric partner, the AhR nuclear translocator (ARNT),84 and further analysis suggested that crosstalk between AhR and Wnt signaling is responsible for the impairment of fin regeneration.86
These studies in zebrafish fin, and a program of endocrine disruptor research in our laboratory, prompted us to re-examine our arrary for members of the AhR-Wnt signaling pathways specifically. Interestingly, a number of these are down-regulated in eye during the early injury phase of optic nerve regeneration (e.g. ahr2, cyp1b1). Although a causal relationship remains to be established, the observations described here illustrate the utility of gene discovery approaches, and comparisons across different test systems, to identify normal regulatory and toxic factors.
Conclusion
In conclusion, these results suggest that the key to regeneration is not the simple addition of growth factors, or the neutralization of inhibitory factors, but rather involvement of many different cell types in creating a permissive environment and a global change in the regulation of apoptosis, proliferation, metabolism, calcium signaling, cell-cell communication and many other cellular functions. Knowledge of gene expression changes in a model of successful regeneration provides an entry point for functional analysis using zebrafish mutants, morpholino knockdown, pharmacological and small molecule applications.
Supplementary Material
Acknowledgments
Research was supported by grants from the National Institutes of Health (NIEHS P42ES07381) and the US Environmental Protection Agency (STAR RD831301), and a traineeship (ATM) from the National Institutes of Health (NICHD 2T 32HD073897). We thank the Boston University Microarray Core Facility for the preparation and processing of the microarrays.
Abbreviations
- PCR
polymerase chain reaction
- RNA
ribonucleic acid
- Ct
cycle threshold
- ANOVA
analysis of variance
- S.E.M
standard error of the mean
- ONX
optic nerve crush
- SHAM
sham surgery
- GO
gene ontology
Footnotes
Authors' Contributions: ATM conceived of the study, has been responsible for all the experimental work, and has been involved in drafting and revising the manuscript. GVC participated as a supervisor in study design and analyses. She has been involved in drafting the manuscript, revising it critically for important intellectual content, and given final approval for the version to be published.
Disclosures: This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
This is an open access article. Unrestricted non-commercial use is permitted provided the original work is properly cited.
References
- 1.Lie DC, Song H, Colamarino SA, Ming GL, Gage FH. Neurogenesis in the adult brain: new strategies for central nervous system diseases. Annu Rev Pharmacol Toxicol. 2004;44:399–421. doi: 10.1146/annurev.pharmtox.44.101802.121631. [DOI] [PubMed] [Google Scholar]
- 2.Benowitz L, Yuqin Y. Rewiring the injured CNS: lessons from the optic nerve. Exp Neurol. 2008;209(2):389–98. doi: 10.1016/j.expneurol.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cajal RY. Degeneration and Regeneration of the Nervous System. New York: Oxford University Press; 1991. [Google Scholar]
- 4.Dezawa M, Adachi-Usami E. Role of Schwann cells in retinal ganglion cell axon regeneration. Prog Retin Eye Res. 2000;19(2):171–204. doi: 10.1016/s1350-9462(99)00010-5. [DOI] [PubMed] [Google Scholar]
- 5.Hirsch S, Bahr M. Growth promoting and inhibitory effects of glial cells in the mammalian nervous system. Adv Exp Med Biol. 1999;468:199–205. doi: 10.1007/978-1-4615-4685-6_16. [DOI] [PubMed] [Google Scholar]
- 6.Lucas SM, Rothwell NJ, Gibson RM. The role of inflammation in CNS injury and disease. Br J Pharmacol. 2006;147(S1):S232–40. doi: 10.1038/sj.bjp.0706400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhatt D, Otto SJ, Depoister B, Fetcho JR. Cyclic AMP-induced repair of zebrafish spinal circuits. Science. 2004;305(5681):254–258. doi: 10.1126/science.1098439. [DOI] [PubMed] [Google Scholar]
- 8.Sperry RW. Patterning of central synapses in regeneration of the optic nerve in teleosts. Physiol Zool. 1948;21:351–61. doi: 10.1086/physzool.21.4.30152014. [DOI] [PubMed] [Google Scholar]
- 9.Bernhardt R, Tongiorgi E, Anzini P, Schachner M. Increased expression of specific recognition molecules by retinal ganglion cells and by optic pathway glia accompanies the successful regeneration of retinal axons in adult zebrafish. J Comp Neurol. 1996;376(2):253–64. doi: 10.1002/(SICI)1096-9861(19961209)376:2<253::AID-CNE7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Lanners HN, Grafstein B. Early stages of axonal regeneration in the goldfish optic tract: an electron microscopic study. J Neurocytol. 1980;9(6):733–51. doi: 10.1007/BF01205016. [DOI] [PubMed] [Google Scholar]
- 11.Attardi DG, Sperry RW. Preferential selection of central pathways by regenerating optic fibers. Exp Neurol. 1963;7:46–64. doi: 10.1016/0014-4886(63)90093-1. [DOI] [PubMed] [Google Scholar]
- 12.Meyer RL. Mapping the normal and regenerating retinotectal projection of goldfish with autoradiographic methods. J Comp Neurol. 1980;189(2):273–89. doi: 10.1002/cne.901890205. [DOI] [PubMed] [Google Scholar]
- 13.Perrone-Bizzozero NI, Benowitz LI. Expression of a 48-kilodalton growth-associated protein in the goldfish retina. J Neurochem. 1987;48(2):644–52. doi: 10.1111/j.1471-4159.1987.tb04141.x. [DOI] [PubMed] [Google Scholar]
- 14.Perry GW, Burmeister DW, Grafstein B. Changes in protein content of goldfish optic nerve during degeneration and regeneration following nerve crush. J Neurochem. 1985;44(4):1142–51. doi: 10.1111/j.1471-4159.1985.tb08736.x. [DOI] [PubMed] [Google Scholar]
- 15.Grafstein B, Murray M. Transport of protein in goldfish optic nerve during regeneration. Exp Neurol. 1969;25(4):494–508. doi: 10.1016/0014-4886(69)90093-4. [DOI] [PubMed] [Google Scholar]
- 16.Quitschke W, Schechter N. In vitro protein synthesis in the goldfish retinotectal pathway during regeneration: evidence for specific axonal proteins of retinal origin in the optic nerve. J Neurochem. 1983;41(4):1137–42. doi: 10.1111/j.1471-4159.1983.tb09063.x. [DOI] [PubMed] [Google Scholar]
- 17.Benowitz LI, Yoon MG, Lewis ER. Transported proteins in the regenerating optic nerve: regulation by interactions with the optic tectum. Science. 1983;222(4620):185–8. doi: 10.1126/science.6194562. [DOI] [PubMed] [Google Scholar]
- 18.Grafstein B. The retina as a regenerating organ. In: Adler R, Farber D, editors. The Retina: A Model for Cell Biology Studies. New York: Academic; 1986. pp. 275–335. [Google Scholar]
- 19.McQuarrie IG, Grafstein B. Protein synthesis and axonal transport in goldfish retinal ganglion cells during regeneration accelerated by a conditioning lesion. Brain Res. 1982;251(1):25–37. doi: 10.1016/0006-8993(82)91270-7. [DOI] [PubMed] [Google Scholar]
- 20.Bormann P, Zumsteg VM, Roth LW, Reinhard E. Target contact regulates GAP-43 and alpha-tubulin mRNA levels in regenerating retinal ganglion cells. J Neurosci Res. 1998;52(4):405–19. doi: 10.1002/(SICI)1097-4547(19980515)52:4<405::AID-JNR4>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 21.Hieber V, Dai X, Foreman M, Goldman D. Induction of alpha1-tubulin gene expression during development and regeneration of the fish central nervous system. J Neurobiol. 1998;37(3):429–40. doi: 10.1002/(sici)1097-4695(19981115)37:3<429::aid-neu8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 22.Becker CG, Becker T. Growth and pathfinding of regenerating axons in the optic projection of adult fish. J Neurosci Res. 2007;85(12):2793–9. doi: 10.1002/jnr.21121. [DOI] [PubMed] [Google Scholar]
- 23.Udvadia AJ, Köster RW, Pate Skene JH. GAP-43 promoter elements in transgenic zebrafish reveal a difference in signals for axon growth during CNS development and regeneration. Development. 2001;128:1175–82. doi: 10.1242/dev.128.7.1175. [DOI] [PubMed] [Google Scholar]
- 24.Becker T, Bernhardt RR, Reinhard E, Wullimann MF, Tongiorgi E, Schachner M. Readiness of zebrafish brain neurons to regenerate a spinal axon correlates with differential expression of specific cell recognition molecules. J Neurosci. 1998;18(15):5789–803. doi: 10.1523/JNEUROSCI.18-15-05789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wanner M, Lang DM, Bandtlow CE, Schwab ME, Bastmeyer M, Stuermer CA. Reevaluation of the growth-permissive substrate properties of goldfish optic nerve myelin and myelin proteins. J Neurosci. 1995;15(11):7500–8. doi: 10.1523/JNEUROSCI.15-11-07500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen E, Ekker SC. Zebrafish as a genomics research model. Curr Pharm Biotechnol. 2004;5(5):409–13. doi: 10.2174/1389201043376652. [DOI] [PubMed] [Google Scholar]
- 27.Cameron D, Gentile KL, Middleton FA, Yurco P. Gene expression profiles of intact and regenerating zebrafish retina. Mol Vis. 2005;11:775–91. [PubMed] [Google Scholar]
- 28.Qin Z, Barthel LK, Raymond PA. Genetic evidence for shared mechanisms of epimorphic regeneration in zebrafish. Proc Natl Acad Sci U S A. 2009;106(23):9310–5. doi: 10.1073/pnas.0811186106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veldman M, Bemben MA, Thompson RC, Goldman D. Gene expression analysis of zebrafish retinal ganglion cells during optic nerve regeneration identifies KLF6a and KLF7a as important regulators of axon regeneration. Dev Biol. 2007;312(2):596–612. doi: 10.1016/j.ydbio.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 30.Lien C, Schebesta M, Makino S, Weber GJ, Keating MT. Gene expression analysis of zebrafish heart regeneration. PLoS Biol. 2006;4(8):260. doi: 10.1371/journal.pbio.0040260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schebesta M, Lien CL, Engel FB, Keating MT. Transcriptional profiling of caudal fin regeneration in zebrafish. Scientific World Journal. 2006;6:38–54. doi: 10.1100/tsw.2006.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westerfield M. The Zebrafish Book 4. Eugene, OR: University of Oregon Press; 2000. [Google Scholar]
- 33.Tchoudakova A, Kishida M, Wood E, Callard GV. Promoter characteristics of two cyp19 genes differentially expressed in the brain and ovary of teleost fish. J Steroid Biochem Mol Biol. 2001;78(5):427–39. doi: 10.1016/s0960-0760(01)00120-0. [DOI] [PubMed] [Google Scholar]
- 34.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCurley AT, Callard GV. Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol Biol. 2008;9:102. doi: 10.1186/1471-2199-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller PY, Janovjak H, Miserez AR, Dobbie Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques. 2002;32(6):1372–4. 6, 8–9. [PubMed] [Google Scholar]
- 37.Chen J, Ruan H, Ng SM, et al. Loss of function of def selectively up-regulates Delta113p53 expression to arrest expansion growth of digestive organs in zebrafish. Genes Dev. 2005;19(23):2900–11. doi: 10.1101/gad.1366405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsukawa T, Arai K, Koriyama Y, Liu Z, Kato S. Axonal regeneration of fish optic nerve after injury. Biol Pharm Bull. 2004;27(4):445. doi: 10.1248/bpb.27.445. [DOI] [PubMed] [Google Scholar]
- 39.Asch W, Leake D, Canger AK, Passini MA, Argenton F, Schechter N. Cloning of zebrafish neurofilament cDNAs for plasticin and gefiltin: increased mRNA expression in ganglion cells after optic nerve injury. J Neurochem. 1998;71(1):20–32. doi: 10.1046/j.1471-4159.1998.71010020.x. [DOI] [PubMed] [Google Scholar]
- 40.Saula KE, Kokea JR, García DM. Activating transcription factor 3 (ATF3) expression in the neural retina and optic nerve of zebrafish during optic nerve regeneration. Comp Biochem Physiol A Mol Integr Physiol. 2010;155(2):172–82. doi: 10.1016/j.cbpa.2009.10.042. [DOI] [PubMed] [Google Scholar]
- 41.Becker CG, Schweitzer J, Feldner J, Schachner M, Becker T. Tenascin-R as a repellent guidance molecule for newly growing and regenerating optic axons in adult zebrafish. Mol Cell Neurosci. 2004;26(3):376–89. doi: 10.1016/j.mcn.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 42.Zhou LX, Wang ZR. Changes in number and distribution of retinal ganglion cells after optic nerve crush in zebrafish. Shi Yan Sheng Wu Xue Bao. 2002;35(2):159–62. [PubMed] [Google Scholar]
- 43.Murray M. A quantitative study of regenerative sprouting by optic axons in goldfish. J Comp Neurol. 1982;209(4):352–62. doi: 10.1002/cne.902090405. [DOI] [PubMed] [Google Scholar]
- 44.Berry M, Rees L, Hall S, Yiu P, Sievers J. Optic axons regenerate into sciatic nerve isografts only in the presence of Schwann cells. Brain Res Bull. 1988;20(2):223–31. doi: 10.1016/0361-9230(88)90182-7. [DOI] [PubMed] [Google Scholar]
- 45.Quigley HA, Nickells RW, Kerrigan LA, Pease ME, Thibault DJ, Zack DJ. Retinal ganglion cell death in experimental glaucoma and after axotomy occurs by apoptosis. Invest Ophthalmol Vis Sci. 1995;36(5):774–86. [PubMed] [Google Scholar]
- 46.Kustermann S, Schmid S, Biehlmaier O, Kohler K. Survival, excitability, and transfection of retinal neurons in an organotypic culture of mature zebrafish retina. Cell Tissue Res. 2008;332(2):195–209. doi: 10.1007/s00441-008-0589-5. [DOI] [PubMed] [Google Scholar]
- 47.Garcia DM, Koke JR. Astrocytes as gate-keepers in optic nerve regeneration— A mini-review. Comp Biochem Physiol A Mol Integr Physiol. 2009;152(2):135–8. doi: 10.1016/j.cbpa.2008.09.026. [DOI] [PubMed] [Google Scholar]
- 48.Stys PK. Anoxic and ischemic injury of myelinated axons in CNS white matter: from mechanistic concepts to therapeutics. J Cereb Blood Flow Metab. 1998;18(1):2–25. doi: 10.1097/00004647-199801000-00002. [DOI] [PubMed] [Google Scholar]
- 49.Xiong Y, Peterson PL, Lee CP. Alterations in cerebral energy metabolism induced by traumatic brain injury. Neurol Res. 2001;23(2–3):129–38. doi: 10.1179/016164101101198460. [DOI] [PubMed] [Google Scholar]
- 50.Verweij BH, Amelink GJ, Muizelaar JP. Current concepts of cerebral oxygen transport and energy metabolism after severe traumatic brain injury. Prog Brain Res. 2007;161:111–24. doi: 10.1016/S0079-6123(06)61008-X. [DOI] [PubMed] [Google Scholar]
- 51.Hertz L. Bioenergetics of cerebral ischemia: a cellular perspective. Neuropharmacology. 2008;55(3):289–309. doi: 10.1016/j.neuropharm.2008.05.023. [DOI] [PubMed] [Google Scholar]
- 52.Aoyama K, Burns DM, Suh SW, et al. Acidosis causes endoplasmic reticulum stress and caspase-12-mediated astrocyte death. J Cereb Blood Flow Metab. 2005;25(3):358–70. doi: 10.1038/sj.jcbfm.9600043. [DOI] [PubMed] [Google Scholar]
- 53.Xing G, Ren M, Watson WA, O'Neil JT, Verma A. Traumatic brain injury-induced expression and phosphorylation of pyruvate dehydrogenase: a mechanism of dysregulated glucose metabolism. Neurosci Lett. 2009;454(1):38–42. doi: 10.1016/j.neulet.2009.01.047. [DOI] [PubMed] [Google Scholar]
- 54.Oertel MF, Schwedler M, Stein M, et al. Cerebral energy failure after subarachnoid hemorrhage: the role of relative hyperglycolysis. J Clin Neurosci. 2007;14(10):948–54. doi: 10.1016/j.jocn.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 55.Weber JT. Calcium homeostasis following traumatic neuronal injury. Curr Neurovasc Res. 2004;1(2):151–71. doi: 10.2174/1567202043480134. [DOI] [PubMed] [Google Scholar]
- 56.Wojda U, Salinska E, Kuznicki J. Calcium ions in neuronal degeneration. IUBMB. 2008;60(9):575–90. doi: 10.1002/iub.91. [DOI] [PubMed] [Google Scholar]
- 57.Vosler PS, Brennan CS, Chen J. Calpain-mediated signaling mechanisms in neuronal injury and neurodegeneration. Mol Neurobiol. 2008;38(1):78–100. doi: 10.1007/s12035-008-8036-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wenk GL. Neuropathologic changes in Alzheimer's disease: potential targets for treatment. J Clin Psychiatry. 2006;67 3:3–7. [PubMed] [Google Scholar]
- 59.Vihtelic TS, Fadool JM, Gao J, Thornton KA, Hyde DR, Wistow G. Expressed sequence tag analysis of zebrafish eye tissues for NEIBank. Mol Vis. 2005;11:1083–100. [PubMed] [Google Scholar]
- 60.Pavlidis M, Fischer D, Thanos S. Photoreceptor degeneration in the RCS rat attenuates dendritic transport and axonal regeneration of ganglion cells. Invest Ophthalmol Vis Sci. 2000;41(8):2318–28. [PubMed] [Google Scholar]
- 61.Nagashima M, Sakuraia H, Mawataria K, Koriyamab Y, Matsukawab T, Katob S. Involvement of retinoic acid signaling in goldfish optic nerve regeneration. Neurochem Int. 2008;54(3–4):229–36. doi: 10.1016/j.neuint.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 62.Goldberg JL, Klassen MP, Hua Y, Barres BA. Amacrine-Signaled Loss of Intrinsic Axon Growth Ability by Retinal Ganglion Cells. Science. 2002;296(5574):1860–4. doi: 10.1126/science.1068428. [DOI] [PubMed] [Google Scholar]
- 63.Shaw PJ, Eggett CJ. Molecular factors underlying selective vulnerability of motor neurons to neurodegeneration in amyotrophic lateral sclerosis. J Neurol. 2000;247 1:I17–27. doi: 10.1007/BF03161151. [DOI] [PubMed] [Google Scholar]
- 64.Reynolds GP, Zhang ZJ, Beasley CL. Neurochemical correlates of cortical GABAergic deficits in schizophrenia: selective losses of calcium binding protein immunoreactivity. Brain Res Bull. 2001;55(5):579–84. doi: 10.1016/s0361-9230(01)00526-3. [DOI] [PubMed] [Google Scholar]
- 65.Fischer D, Hauk TG, Müller A, Thanos S. Crystallins of the beta/gamma-superfamily mimic the effects of lens injury and promote axon regeneration. Mol Cell Neurosci. 2008;37(3):471–9. doi: 10.1016/j.mcn.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 66.Filbin MT. How inflammation promotes regeneration. Nat Neurosci. 2006;9(6):715–7. doi: 10.1038/nn0606-715. [DOI] [PubMed] [Google Scholar]
- 67.Leon S, Yin Y, Nguyen J, Irwin N, Benowitz LI. Lens injury stimulates axon regeneration in the mature rat optic nerve. J Neurosci. 2000;20(12):4615–26. doi: 10.1523/JNEUROSCI.20-12-04615.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cui Q, Yin Y, Benowitz LI. The role of macrophages in optic nerve regeneration. Neuroscience. 2009;158(3):1039–48. doi: 10.1016/j.neuroscience.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sandvig A, Berry M, Barrett LB, Butt A, Logan A. Myelin-, reactive glia-, and scar-derived CNS axon growth inhibitors: expression, receptor signaling, and correlation with axon regeneration. Glia. 2004;46(3):225–51. doi: 10.1002/glia.10315. [DOI] [PubMed] [Google Scholar]
- 70.Löw K, Culbertson M, Bradke F, Tessier-Lavigne M, Tuszynski MH. Netrin-1 is a novel myelin-associated inhibitor to axon growth. J Neurosci. 2008;28(5):1099–108. doi: 10.1523/JNEUROSCI.4906-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berry M, Ahmed Z, Lorber B, Douglas M, Logan A. Regeneration of axons in the visual system. Restor Neurol Neurosci. 2008;26(2–3):147–74. [PubMed] [Google Scholar]
- 72.Liberto CM, Albrecht PJ, Herx LM, Yong VW, Levison SW. Pro- regenerative properties of cytokine-activated astrocytes. J Neurochem. 2004;89(5):1092–100. doi: 10.1111/j.1471-4159.2004.02420.x. [DOI] [PubMed] [Google Scholar]
- 73.Nakase T, Söhl G, Theis M, Willecke K, Naus CC. Increased apoptosis and inflammation after focal brain ischemia in mice lacking connexin43 in astrocytes. Am J Pathol. 2004;164(6):2067–75. doi: 10.1016/S0002-9440(10)63765-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lagrèze WA, Pielen A, Steingart R, et al. The peptides ADNF-9 and NAP increase survival and neurite outgrowth of rat retinal ganglion cells in vitro. Invest Ophthalmol Vis Sci. 2005;46(3):933–8. doi: 10.1167/iovs.04-0766. [DOI] [PubMed] [Google Scholar]
- 75.Wong WK, Cheung AW, Cho EY. Lens epithelial cells promote re-growth of retinal ganglion cells in culture and in vivo. Neuroreport. 2006;17(7):699–704. doi: 10.1097/01.wnr.0000215776.91850.96. [DOI] [PubMed] [Google Scholar]
- 76.Inatani M, Honjo M, Otori Y, et al. Inhibitory effects of neurocan and phosphacan on neurite outgrowth from retinal ganglion cells in culture. Invest Ophthalmol Vis Sci. 2001;42(8):1930–8. [PubMed] [Google Scholar]
- 77.Sajnani G, Aricescu AR, Jones EY, Gallagher J, Alete D, Stoker A. PTPsigma promotes retinal neurite outgrowth non-cell-autonomously. J Neurobiol. 2005;65(1):59–71. doi: 10.1002/neu.20175. [DOI] [PubMed] [Google Scholar]
- 78.Eitan S, Solomon A, Lavie V, et al. Recovery of visual response of injured adult rat optic nerves treated with transglutaminase. Science. 1994;264(5166):1764–8. doi: 10.1126/science.7911602. [DOI] [PubMed] [Google Scholar]
- 79.Kassen SC, Ramanan V, Montgomery JE, et al. Time course analysis of gene expression during light-induced photoreceptor cell death and regeneration in albino zebrafish. Dev Neurobiol. 2007;67(8):1009–31. doi: 10.1002/dneu.20362. [DOI] [PubMed] [Google Scholar]
- 80.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298(5601):2188–90. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 81.Poss KD, Keating MT, Nechiporuk A. Tales of regeneration in zebrafish. Dev Dyn. 2003;226(2):202–10. doi: 10.1002/dvdy.10220. [DOI] [PubMed] [Google Scholar]
- 82.Scott IC, Stainier DY. Development. Fishing out a new heart. Science. 2002;298(5601):2141–2. doi: 10.1126/science.1079821. [DOI] [PubMed] [Google Scholar]
- 83.Zodrow J, Tanguay RL. 2,3,7,8-tetrachlorodibenzo-p-dioxin inhibits zebrafish caudal fin regeneration. Toxicol Sci. 2003;76(1):151–61. doi: 10.1093/toxsci/kfg205. [DOI] [PubMed] [Google Scholar]
- 84.Mathew LK, Andreasen EA, Tanguay RL. Aryl hydrocarbon receptor activation inhibits regenerative growth. Mol Pharmacol. 2006;69(1):257–65. doi: 10.1124/mol.105.018044. [DOI] [PubMed] [Google Scholar]
- 85.Mitchell KA, Lockhart CA, Huang G, Elferink CJ. Sustained aryl hydrocarbon receptor activity attenuates liver regeneration. Mol Pharmacol. 2006;70(1):163–70. doi: 10.1124/mol.106.023465. [DOI] [PubMed] [Google Scholar]
- 86.Mathew LK, Sengupta SS, Ladu J, Andreasen EA, Tanguay RL. Crosstalk between AHR and Wnt signaling through R-Spondin1 impairs tissue regeneration in zebrafish. FASEB J. 2008;22(8):3087–96. doi: 10.1096/fj.08-109009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.