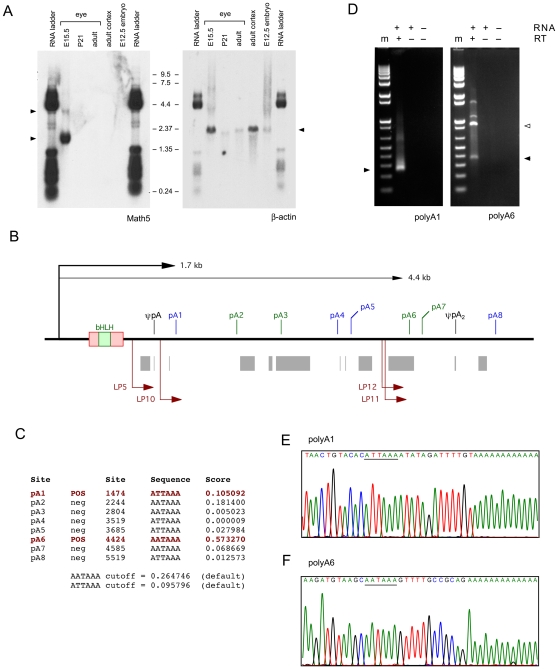
Figure 2. Math5 messenger RNAs.
A. Northern blot probed with 1.2 kb Math5 (JN4C) and 1.1 kb β-actin cDNAs. Two Math5 mRNAs are visible (left arrowheads), but no hybridizing RNA species is present in the 0.8–1.0 kb size range. The RNA size ladder cross-hybridized to vector DNA in the plasmid probes. B. Map of the 3′UTR and flanking genomic DNA (6 kb), showing eight potential polyA signals ATTAAA (blue) and AATAAA (green); the internal A14 priming site in the UTR (ψpA); interspersed repeats (gray); and the nested 3′RACE primers (dark red) for pA1 and pA6 sites, which have the most favorable sequence context. Clones JN2 and BC092234 terminate at pA1, whereas cDNAs JN1, JN4 and JN6 terminate at ψpA [7], [13]. ψpA2 marks an A-rich genomic site captured in the pA6 assay. C. polyADQ scores for all potential pA sites, calculated using human genome parameters [22]. Only pA1 and pA6 have scores above threshold. D. Embryonic eye RT-PCRs with 260 bp and 365 bp 3′RACE products (arrowheads) showing utilization of pA1 and pA6 sites. The 900 bp product was primed from ψpA2 (open arrowhead). m, marker (1 kb-plus ladder); RT, reverse transcriptase. E. Sequence of pA1 RACE products originating from the 1.7 kb Math5 mRNA. F. Sequence of pA6 RACE products originating from the 4.4 kb Math5 mRNA.