Abstract
Forest trees are ideally suited to association mapping due to their high levels of diversity and low genomic linkage disequilibrium. Using an association mapping approach, single-nucleotide polymorphism (SNP) markers influencing quantitative variation in wood quality were identified in a natural population of Pinus radiata. Of 149 sites examined, 10 demonstrated significant associations (P < 0.05, q < 0.1) with one or more traits after accounting for population structure and experimentwise error. Without accounting for marker interactions, phenotypic variation attributed to individual SNPs ranged from 2 to 6.5%. Undesirable negative correlations between wood quality and growth were not observed, indicating potential to break negative correlations by selecting for individual SNPs in breeding programs. Markers that yielded significant associations were reexamined in an Australian land race. SNPs from three genes (PAL1, PCBER, and SUSY) yielded significant associations. Importantly, associations with two of these genes validated associations with density previously observed in the discovery population. In both cases, decreased wood density was associated with the minor allele, suggesting that these SNPs may be under weak negative purifying selection for density in the natural populations. These results demonstrate the utility of LD mapping to detect associations, even when the power to detect SNPs with small effect is anticipated to be low.
NUMEROUS traits of agronomic importance are demonstrated to be under genetic control (Keurentjes et al. 2008), and there is considerable interest in characterizing the causative polymorphisms underlying their quantitative variation. In forest trees, quantitative variation in traits such as mechanical and pulping properties of wood, growth, cold hardiness, and drought acclimation are likely to result from allelic variation within multiple genes (Neale and Savolainen 2004; Oraguzie and Wilcox 2007). Because of the commercial importance of radiata pine (Pinus radiata D. Don), we are exploring the molecular basis of variation in its wood properties using an association genetics approach.
Radiata pine wood properties are variable in domesticated populations and exhibit a quantitative mode of inheritance with high heritability, indicating a strong underlying genetic component. High heritabilities have been observed for solid wood traits including density, cellulose microfibril angle (MFA), and modulus of elasticy (MOE) (Baltunis et al. 2007) and for carbohydrate composition, pulp yield, fiber length, and perimeter (Evans et al. 1997; Kibblewhite 1999); and genetic control has been established for several important production traits (Kumar 2004; Dungey et al. 2006; Gapare et al. 2006; Wu et al. 2007).
It is anticipated that variation in wood quality will be dependent on variation in numerous genes involved in xylogenesis. Studies of gene expression in developing xylem have revealed genes involved in spatially and temporally regulated processes such as cambial division, cell differentiation, expansion, and secondary cell wall biosynthesis (Allona et al. 1998; Paux et al. 2005; Pavy et al. 2005; Cato et al. 2006; Li et al. 2009). Studies in model plants indicate several thousand genes are likely to be involved in these processes, which can be grouped into four categories: information storage and processing, cellular processes and signaling, metabolism, and genes with unknown functions (Yuan et al. 2007). Studies in model plants have revealed specific functions for numerous genes in these developmental pathways. To date there have been few functional studies in forest trees revealing genes directly affecting wood quality (MacKay et al. 1997; Tsai et al. 1998; Valerio et al. 2003; Lu et al. 2004; Spokevicius et al. 2007), largely due to inherent features of forest trees including their long generation times, large size, and lack of inbred or pure lines (Oh et al. 2003).
Attempts to identify genes underlying phenotypic variation in forest trees typically focused on quantitative trait loci (QTL) mapping. In radiata pine several QTL associated with wood density, growth, and disease response have been identified (Devey et al. 2004a,b; Matheson et al. 2006). However, poor success in validating QTL markers in populations with different genetic and environmental backgrounds due to unfavorable pedigree linkage disequilibrium (LD) (Neale and Savolainen 2004), has led to a shift in research to population-level association (or LD) mapping. This approach aims to identify gene alleles that directly contribute to phenotypic variation by statistical inference of the cosegregation of genotype and phenotype data in unrelated populations (Neale and Savolainen 2004). Forest trees are ideally suited to association mapping as LD and population structure are frequently low (Neale and Savolainen 2004). Encouraging progress toward identifying molecular polymorphisms linked to phenotypic variation in trees has recently been achieved following this approach. The cinnamoyl CoA reductase gene in Eucalyptus (Thumma et al. 2005) and four cell wall genes in P. taeda (Gonzalez-Martinez et al. 2007) were shown to influence variation in MFA, early wood specific gravity, and late wood proportion. Recently a single-nucleotide polymorphism (SNP) in a cobra-like gene in Eucalyptus nitens was identified that is associated with cellulose content and pulp yield (Thumma et al. 2009). Further functional analysis revealed that this SNP is a regulatory polymorphism and is likely to affect both traits through its influence on allelic expression. This finding demonstrates the utility of populations harboring low LD in the study of causative polymorphisms. Association genetics has also been applied in model plants to identify SNPs influencing major traits in Arabidopsis and maize (Thornsberry et al. 2001; Olsen et al. 2004; Aranzana et al. 2005) and has been used extensively to dissect complex traits in humans where population structure parallels that of many tree species (Pritchard and Przeworski 2001; Burton et al. 2007).
Despite the potential promised from association genetics, several factors have been identified that influence power to detect true associations (Long and Langley 1999; Neale and Savolainen 2004; Gordon and Finch 2005; Newton-Cheh and Hirschhorn 2005). These factors can often be accounted for at the experimental design phase, by considering the extent of genome-wide LD, the candidate genes selected, the effect size, the number of genes affecting the trait, allele frequency, sample size, and rates of genotyping and phenotyping error. The use of natural populations or pedigrees for association studies can introduce confounding genetic structure (Burton et al. 2007; Zhao et al. 2007), which creates false LD between markers and QTL (Neale and Savolainen 2004; Ball 2007). Radiata pine occurs naturally as a regional ensemble of disjoint populations over a small range including three from mainland California and two on Guadalupe and Cedros Islands. Genetic diversity of the mainland populations, used in this study, is at least moderate among conifers (Karhu et al. 2006), and the populations exhibit a low yet significant level of genetic structure (FST ∼ 5%) (Plessas and Strauss 1986; Karhu et al. 2006), which may confound association tests. The aim of this study was to apply an LD mapping approach to identify allelic variants regulating wood formation in the natural populations of radiata pine. Several methods have been developed for the quantification of genetic structure and we have employed two of these to correct for allelic structure arising in populations and pedigrees when testing individual SNP associations (Ritland 1996; Pritchard et al. 2000). Because of the large size, anonymity, and complexity of conifer genomes (12n = 26,000 Mb) (Ahuja and Neale 2005), and the rapid breakdown of LD, we focused on SNPs from 36 cell wall candidate genes and their flanking sequences rather than a genome-wide scan. Where SNP associations were detected in the natural populations, we investigated whether significant associations could be validated in an Australia land race.
MATERIALS AND METHODS
Genetic material:
P. radiata trees in the discovery population were maintained in a 25-year-old provenance-progeny trial at Batlow, New South Wales (35°31′17″S; 148°08′40″E). This trial was established following an incomplete block design, with four trees per family per replicate, from seed collected from the species natural distribution in mainland California. Needles were sampled from one tree per family for 447 families in replicate 1 and included all three mainland provenances: Monterey (210), Año Nuevo (155), and Cambria (82). Needles were also collected from a second-generation progeny trial in Flynn, Victoria (BR9611: 38°15′46″S; 146°41′33″E), which served as a validation population that was previously described by Baltunis et al. (2007) and is composed of families descended from the Año Nuevo and Monterey provenances. This population was part of a 7-year-old trial established following an incomplete block design with four trees per family per replicate. Needles were collected from 458 individuals. This population contained two half-sibs (open pollinated) from each of 229 different families. Diploid DNA was extracted from needles using a modified CTAB protocol (Doyle and Doyle 1990) and further purified on QIAquick PCR purification columns according to the manufacturer's instructions.
Candidate gene selection and SNP genotyping:
Thirty-eight candidate genes chosen on the basis of their known or likely involvement in development of secondary xylem were previously sequenced in a panel of radiata pine individuals (Table 1). This included genes in five functional categories: (1) lignin biosynthesis, (2) cellulose synthesis, (3) cell wall structure, (4) cell expansion, and (5) abiotic stress responses. Prior to genotyping, SNPs were parsed from 72 candidate gene amplicon alignments covering 54,188 bp of DNA sequence (supporting information, Table S1). One hundred forty-nine haplotype-tagging SNPs were genotyped in parallel across the 447 individuals from the discovery population, using universal bead arrays (Illumina) (Shen et al. 2005). Linkage disequilibrium between unphased SNP data was assessed using HelixTree software 6.3.6 (Golden Helix). The overall level of LD between SNPs was low with <1.2% of pairwise correlations between sites (r2) exceeding 0.2 (Table S2). Significant SNP associations identified in the discovery population were regenotyped in 458 individuals from the validation population, using the iPLEX Gold assay (Sequenom).
TABLE 1.
Distribution of SNP markers across 36 candidate genes sequenced in the Batlow discovery population
| Gene | Name | Amplicons | L | SNPs |
|---|---|---|---|---|
| α-Expansin | αexp1 | 4 | 1,492 | 7 |
| α-Tubulin | αtub | 1 | 788 | 2 |
| Aquaporin | porin | 1 | 1,919 | 5 |
| COBL4 (Arabidopsis thaliana) | cobl4 | 2 | 2,642 | 4 |
| Dehydrin 2 (A. thaliana) | dh2 | 1 | 606 | 2 |
| Dehydrin 7/8 (A. thaliana) | dh7/8 | 1 | 232 | 2 |
| rac13 GTPase (G. hirsutism) | rac13 | 4 | 3,372 | 6 |
| Caffeate 3-O-methyltransferase 2 (P. taeda) | comt2 | 2 | 1,290 | 2 |
| Chloroplastic superoxide dismutase | sodchl | 1 | 720 | 3 |
| Cinnamate 4-hydroxylase | c4h | 1 | 440 | 2 |
| Cinnamoyl-CoA reductase | ccr1 | 2 | 2,763 | 2 |
| Cinnamyl-alcohol dehydrogenase | cad | 3 | 2,111 | 11 |
| Korrigan (endo-1,4-b-D-glucanase) | kor | 3 | 2,523 | 5 |
| Katanin (fragile fiber mutant 2) (A. thaliana) | fra2 | 1 | 786 | 3 |
| Glycine hydroxymethyltransferase | glyhmt | 2 | 1,300 | 5 |
| hdzip transcription factor | athb8 | 2 | 1,621 | 7 |
| hdzip transcription factor | athbx | 1 | 470 | 2 |
| Peroxidase precursor | perox | 1 | 812 | 1 |
| Phenylcoumaran benzylic ether reductase | pcber | 1 | 572 | 3 |
| Proline-rich protein | prp | 1 | 643 | 6 |
| Cellulose synthase 3 (P. radiata) | cesa1 | 10 | 7,649 | 19 |
| Cellulose synthase 7 (P. radiata) | cesa2 | 1 | 716 | 1 |
| Cellulose synthase 1 (P. radiata) | cesa3 | 3 | 2,244 | 6 |
| lim 1 transcription factor (P. taeda) | lim1 | 2 | 1,702 | 2 |
| myb 1 transcription factor (P. taeda) | myb1 | 1 | 1,361 | 2 |
| myb 4 transcription factor (P. taeda) | myb4 | 2 | 1,256 | 2 |
| Phenylalanine ammonia lyase 1 (P. taeda) | pal 1 | 3 | 2,128 | 6 |
| Arabinogalactan protein 4 (P. taeda) | agp4 | 2 | 1,749 | 6 |
| Arabinogalactan protein 6 (P. taeda) | agp6 | 1 | 497 | 1 |
| Glycine-rich cell wall protein | lp5 | 1 | 735 | 1 |
| S-adenosyl-L-homocysteine hydrolase | sahh1 | 1 | 650 | 3 |
| S-adenosyl-L-methionine synthase 1 (P. taeda) | sam1 | 2 | 1,342 | 6 |
| S-adenosyl-L-methionine synthase 2 (P. taeda) | sam2 | 2 | 1,399 | 6 |
| Sucrose synthase | susy | 2 | 1,043 | 2 |
| Unknown protein | nh3702 | 1 | 287 | 1 |
| Xyloglucan endotransglycosylase 1 (P. taeda) | xet1 | 3 | 2,328 | 5 |
| Total | 54,188 | 149 |
L, length of sequenced region.
Trait data analysis:
Discovery population:
Increment cores were sampled at breast height from the north side of each tree in the discovery population. Cell wall dimensions and wood quality data were collected via X-ray diffraction and point measurement on 1.5 × 0.3-cm wooden strips prepared from 12-mm incremental cores using SilviScan. Nine traits were measured including density, MOE, MFA, fiber wall thickness, fiber wall tangential diameter, fiber wall radial diameter, fiber coarseness, fiber-specific surface area, and cell population.
The first cambial ring does not necessarily correspond to the same calendar year tree to tree, and therefore increment cores were aligned between samples on the basis of cambial ring widths. On average, 20.5 rings were simultaneously partitioned along the core for each trait on the basis of individual density profiles, using Analyze (Maddock 2001). This process yielded ring-by-ring early and late wood estimates for each trait, as well as late wood proportion. Transition age was estimated for each sample using established methods (Gapare et al. 2006). Trait data were subsequently separated into juvenile and mature phases, where juvenile rings accounted for all rings up to and including the predicted transition ring. Significant differences in wood quality traits as well as gene expression have been reported between the juvenile and mature developmental stages (Li et al. 2009), prompting their analysis as separate components in addition to whole-core estimates.
Three principal components describing the uncorrelated variation present among a set of 13 whole-core traits (density, MFA, MOE, fiber wall thickness, fiber wall tangential diameter, fiber wall radial diameter, fiber coarseness, fiber-specific surface area, cell population, minimum density, maximum density, ring width, and cumulating ring width) were defined using StatistiXL 1.7 software and an eigenvalue cutoff of 1. The final trait set used for association analysis in the discovery population consisted of 62 directly measured and synthetic (including principal components) traits, which are listed in Table S3.
Validation population:
Phenotypic data for core length, density, MOE, and MFA, measured using SilviScan on 12-mm increment cores collected at breast height, were taken from the data set of Baltunis et al. (2007). We selected ring-by-ring and whole-core averages for the outer four of seven rings for each trait, corresponding to the juvenile wood. In total 36 traits were used for association tests (Table S3).
Association analysis:
Discovery population:
Associations between 62 traits and 149 SNPs were tested via a least-squares fixed effects general linear model implemented in Tassel (Bradbury et al. 2007). The statistical model is described by y = Xβ + e, where y is a vector for the observed dependent variable (trait), β is a vector containing independent fixed effects, including genetic marker and population structure matrices, X is the known design matrix, and e is the unobserved vector for the random residual (error) (Henderson 1975). Tests for significance were performed on 447 individuals from the discovery population, Año Nuevo (155), Monterey (210), and Cambria (82), as well as separately within the three natural populations. Since significant divergence has been detected between the three provenances in this population (Karhu et al. 2006), a Q-matrix describing individual population assignments was generated in Structure (Pritchard et al. 2000) and incorporated in the model. We utilized the same 149 SNP markers and 447 samples from the discovery population, assuming independence of markers and applying a previously optimized burn-in of 10,000, and 100,000 chains to estimate likelihoods from MCMC analysis. All P-values were adjusted for multiple testing on the basis of 2000 permutations of each trait across 149 SNP markers. Pedigree structure, or kinship, in the provenance-progeny trial was found to be low following analysis of pairwise kinship coefficients among the same 447 trees and 149 SNPS in Spagedi (Hardy and Vekemans 2002). The Loiselle and Ritland estimators (Loiselle et al. 1995; Ritland 1996) yielded similar results with coefficients ≤0 for a majority of analysis. Associations were tested using the Loiselle matrix and mixed linear model implemented in Tassel (y = Xβ + Zu + e, where X and Z are the known design matrices and u is an unknown vector of random additive genetic effects from multiple-background QTL). Consistent with a lack of kinship, the results were >99% similar to those of the general linear model (Table S4) and were subsequently omitted.
Validation population:
A panel of 25 SNPs producing significant associations in the discovery population using the general linear model were genotyped in the validation population. This included all SNPs shortlisted in Table 3 and 15 SNP markers that had P-values <0.05 in the discovery population but had q-values >0.1 (Table S1). Associations with 36 traits were tested using the same methods as in the discovery population. A Q-matrix was included in the model since significant population genetic structure was identified using Structure (Figure S1). Genotype data for four SNPs (2, 133, 17, and 9) could not be analyzed in the validation population: SNP 2, a low-frequency SNP in the Año Nuevo and Monterey provenances, did not segregate in the validation population; and SNPs 9, 17, and 133 failed quality control. The Loiselle and Ritland estimators yielded pairwise kinship coefficients ≤ 0 >50% of the time, and results for the mixed linear model were >96% similar to those of the general linear model (Table S5). Consequently results for the mixed model were omitted.
TABLE 3.
Shortlisted SNP assocaitions identified in the Batlow association population
| SNP | Gene | Trait | Description | Genotype | Minor allele | P-value | q-value | FST | Bayes factor | Año Nuevoa | Cambriaa | Montereya | RSQ | % trait |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2b | rac13 | Trait 41 | MFA | g/c | 0.072 | 0.0015 | 0.0721 | 0.303 | 33.96 | — | 0.041 | — | 6.5 | 12.8 |
| 2b | rac13 | Trait 50 | MFA | g/c | 0.072 | 0.0035 | 0.0888 | 0.303 | 33.96 | — | 0.026 | — | 6.3 | 13.3 |
| 2b | rac13 | Trait 52 | MFA | g/c | 0.072 | 0.0055 | 0.0888 | 0.303 | 16.96 | — | 0.043 | — | 6.4 | 14.7 |
| 11b | porin | PCA2 | PCA2 | g/a | 0.053 | 0.0025 | 0.0721 | 0.038 | 33.98 | 0.006 | — | 0.013 | 3.1 | - |
| 11b | porin | Trait 35 | Coarseness | g/a | 0.053 | 0.0045 | 0.0888 | 0.038 | 25.43 | 0.017 | — | 0.008 | 3.1 | 13.3 |
| 45c | comt | Trait 53 | MOE | g/a | 0.219 | 0.0065 | 0.0888 | 0.001 | 12.35 | — | 0.006 | — | 2.8 | 16.3 |
| 50b | αexpan | Trait 18 | Wall thickness | g/t | 0.367 | 0.0025 | 0.0721 | 0.006 | 10.04 | — | 0.054 | — | 3 | 2.2 |
| 50b | αexpan | Trait 37 | Wall thickness | g/t | 0.367 | 0.0045 | 0.0888 | 0.006 | 10.04 | — | 0.028 | — | 2.7 | 3.1 |
| 57b | lp5 | Trait 18 | Wall thickness | a/g | 0.183 | 0.0020 | 0.0721 | 0.035 | 10.02 | 0.002 | — | 0.013 | 3.2 | 4.6 |
| 57b | lp5 | Trait 37 | Wall thickness | a/g | 0.183 | 0.0055 | 0.0888 | 0.035 | 7.50 | 0.004 | — | 0.014 | 3.6 | 1.1 |
| 57b | lp5 | Trait 39 | Specific surface area | a/g | 0.183 | 0.0065 | 0.0888 | 0.035 | 5.99 | 0.004 | — | 0.003 | 3.7 | 4.9 |
| 57b | lp5 | Trait 19 | Specific surface area | a/g | 0.183 | 0.0075 | 0.0927 | 0.035 | 5.99 | 0.003 | — | 0.008 | 3.3 | 4.1 |
| 60b | pal1 | Trait 10 | Density | c/t | 0.141 | 0.0005 | 0.0433 | 0.064 | 42.02 | — | — | 0.007 | 2 | 6.7 |
| 69b | athb8 | Trait 32 | Cell population | c/g | 0.139 | 0.0080 | 0.0944 | 0.012 | 88.12 | — | — | 0.083 | 2.7 | 11.5 |
| 84b | cesa3 | Trait 59 | No. juvenile rings | t/c | 0.016 | 0.0075 | 0.0927 | 0.111 | 7.32 | — | 0.013 | — | 2.3 | 16.1 |
| 118b | rac13 | Trait 16 | Cell population | a/g | 0.091 | 0.0065 | 0.0888 | 0.091 | 23.62 | 0.048 | — | 0.012 | 2.8 | 0.7 |
| 118b | rac13 | Trait 33 | Cell population | a/g | 0.091 | 0.0060 | 0.0888 | 0.091 | 14.12 | 0.04 | — | 0.01 | 2.9 | 15.2 |
| 131b | sahh | PCA3 | PCA3 | c/t | 0.107 | 0.0015 | 0.0721 | 0.068 | 60.09 | 0.062 | — | — | 3.1 | — |
RSQ, proportion of the phenotypic variance attributed to SNP; % trait, percentage change in trait mean associated with SNP.
P-values.
Silent SNP.
Nonsynonomous SNP.
An alternative statistic based on the false discovery rate, or q-value, which provides an additional measure of significance dependent on the P-value distribution, was applied in both populations (Storey and Tibshirani 2003). Individual association q-values were calculated using Q-value software upon P-values generated using the general linear model. Bin counts between P-values of 0.85 and 1 were set to the mean count observed over the remaining distribution to eliminate bias due to the permutation test, which adjusted many P-values to near 1. Bayes factors were calculated for individual SNP associations, using the Bayesian statistical package ldDesign (Ball 2005). The interpretive scale of Kass and Raftery (1995) was used to grade strength of evidence for each association on the basis of estimated Bayes factors, where Bayes factor categories 0–2, 2–6, 6–10, and >10 corresponded to weak, positive, strong, and very strong evidence, respectively. Bayes factor calculation incorporated priors for allele frequency, P-value, and population size.
RESULTS
SNP genotypes:
The quality of GoldenGate genotype scores for individual SNPs was assessed from their GenTrain cluster and GenCall genotype scores in BeadStudio (Illumina). The average GenTrain score across all SNP loci and individuals was 0.67, but ranged from 0.3 to 0.89 for individual SNP clusters. In the most extreme cases the heterozygote cluster tended to be very diffuse, possibly representing variable probe binding efficiency due to anonymous variations in the SNP flanking region. GenCall scores for individual genotypes fell in a similar range, with an average score of 0.65 over all SNPs and individuals and average minimum and maximum scores of 0.25 and 0.89, respectively. This indicates that individual genotype data for the 149 SNPs assayed were of moderate to high quality (i.e., >0.2) and could be included in further analysis. Assessment of allelic segregation among all populations revealed that ∼73% of SNPs adhered to Hardy–Weinberg expectations when P-values were relaxed (<0.10). In some cases departure from the theoretical expectation may have resulted from a Wahlund effect for structured loci. When markers were examined within provenances, 95% adhered to the Hardy–Weinberg expectations in all three populations.
Wood quality trait analysis:
Whole-core traits were shown to be distributed normally following the one-sample Kolmigrov–Smirnov test (P < 0.05). The coefficient of variation was consistently higher for traits measured on the juvenile wood (Figure S2). In particular, juvenile variances for MOE and MFA were high compared to the mature component. Estimation of Pearson's correlation coefficient confirmed strong negative and positive correlations for many of the whole-core traits (Table S6). In general, fiber dimensions were strongly correlated with density and MOE. While MFA and MOE were strongly positively correlated, MFA was in general only moderately correlated with the other traits. The three principal components identified explained 84% of the total trait variance in the data. Graphical distribution of the casewise principal components analysis (PCA) scores for individuals and component score coefficients for traits are presented in Figure S3. Component loadings indicated the major traits contributing to components identified were as follows: PCA1, density, early wood density, MOE, late wood density, wall thickness, cell population, and specific surface area; PCA2, cell population, specific surface area, radial diameter, wall thickness, and coarseness; and PCA3, MFA.
SNPs affecting wood quality:
Significant associations in the discovery population, with and without adjustment for population genetic structure, are summarized in Table 2 and Table S7. In total 47 associations were found to be significant after correction for population structure and multiple testing, or 0.51% of all tests. The cumulative P-value distribution between 0 and 1 plotted as a density histogram highlights the skew in P-values near zero compared to the distribution expected if all SNPs were null (Figure 1). This resulted in significant q-values (<0.1) for approximately half of these associations. This approach was used to assemble a shortlist of 18 SNP associations with 10 SNPs, from nine genes (Table 3). Bayes factors estimated for shortlisted associations indicated very strong evidence for the observed associations as Bayes factors fell between 10 and 88 in all but four instances. The markers identified controlled between 2 and 6.5% of phenotypic variation (Table 3). Not accounting for colinearity between markers, the cumulative percentage of improvement in phenotype ranged between 7 and 41% or for individual traits between 0.7 and 16.3%. Undesirable negative correlations between shortlisted SNP associations and growth were not observed.
TABLE 2.
Summary of association test results for the general linear model in the discovery (Batlow) and validation (Flynn) populations
| No. significant associations |
% significant |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Model | Population | Structure | Permutations | P < 0.001 | P < 0.01 | P < 0.05 | Tests | P < 0.001 | P < 0.01 | P < 0.05 |
| General linear | Discovery | — | 2000 | 16 | 29 | 56 | 9238 | 0.17 | 0.31 | 0.61 |
| General linear | Discovery | Q-matrix | 2000 | 3 | 23 | 47 | 9238 | 0.03 | 0.25 | 0.51 |
| General linear | Validation | — | 2000 | 2 | 6 | 14 | 814 | 0.25 | 0.74 | 1.72 |
| General linear | Validation | Q-matrix | 2000 | 1 | 4 | 12 | 814 | 0.12 | 0.49 | 1.47 |
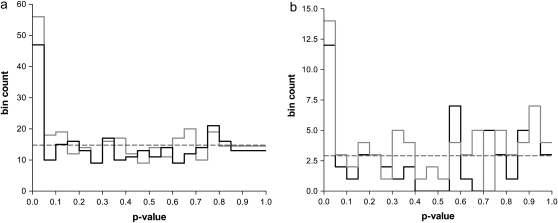
Figure 1.—
(a) P-value density histograms for associations tested in the discovery population (a) and the validation population (b) under the general linear model, with adjustment for structure and multiple testing (solid line). P-values for the general linear model without adjustment for structure are provided for comparison (shaded line). Modal bins were observed in both populations at 0 < P < 0.05, demonstrating a skew in P-values near zero. The dashed line is the density histogram expected if all SNPs were null.
Inclusion of the Q-matrix in the general linear model resulted in a 19% decrease in the number of significant associations at P < 0.05 (Table 2, Figure 1), presumably due to adjusted significance of associations where structure was detected. Genetic differentiation (FST) across the three provenances for individual SNPs ranged between 0.01 and 36.86%, with a number of outliers suggested from the FST histogram (Table 3, Figure S4). The average differentiation among SNPs that dropped out after adjusting for structure was higher (10%) than that in the remaining loci (4%) (ANOVA P = 0.0004). Six SNPs (2, 57, 61, 64, 113, and 133) in the former class retained one or more significant associations, two of which were shortlisted in Table 3. It is possible that the Q-matrix was unable to sufficiently adjust for strong allelic structure in these cases. To explore the validity of these associations we examined the outcome of the model when SNPs were tested in isolation from structure (within populations). Despite small population sizes (n = 82–210), associations for most SNPs in Table 3 remained significant (P ∼ 0.05), including all of the “structured” SNP associations, in one or more of the three populations. This suggests that divergent allele frequencies may have arisen due to population-dependent selection for the trait of interest.
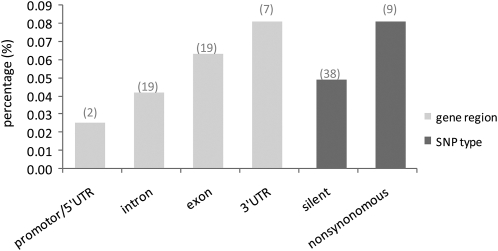
Trends in the frequency of associations possibly due to dependence on SNP function were also observed. When grouped according to physical position (i.e., intron, exon, and 5′- and 3′-UTR), the proportion of associations with P < 0.05 was significantly higher for the 3′-UTR and the exon than for other regions (Figure 2). This fits with the general expectation that polymorphisms from functional regions would more frequently afford phenotypic changes detectable in association tests. In addition, silent polymorphisms yielded 40% fewer significant associations than nonsynonymous sites, indicating a strong bias toward SNP associations where an amino acid has been substituted.
Figure 2.—
Percentage of significant associations (P < 0.05) per gene region and SNP type. Differences in the proportion of significant associations for silent and nonsynonymous sites were significant (P < 0.01) (Z = 2.6) for the binomial test.
SNP validation:
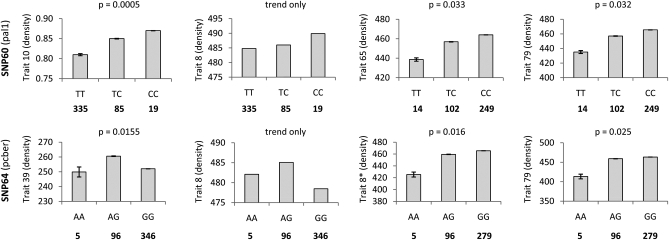
The proportion of significant associations identified in the validation population with and without adjustment for population genetic structure is presented in Table 2. In total 12 associations, with three SNPs, were found to be significant after corrections for population structure and multiple testing were applied, or 1.47% of all tests. This is three times higher than for the discovery population. The cumulative P-value distribution was similarly skewed toward zero (Figure 1), resulting in several associations with significant or near significant q-values (Table 4). Trait means and standard errors categorized by genotype for two of these SNPs are presented in Figure 3, and box-and-whisker plots are presented in Figure S5. For one polymorphism (SNP 60) associations were validated directly with density, meaning that the SNP was significantly associated with this trait type in both populations. The allele effect was conserved between populations, with the TT and CC homozygotes conferring lower and higher trait values, respectively, across the board, with the heterozygote intermediate to these states (Figure 3). Although SNP 60 was not associated with average density (trait 8) in the discovery population, the trend across states was the same for this trait. Reduced power due to sample size and inability to adjust for within-site environmental variation may have affected the sensitivity of this test to detect associations with average density in the discovery population. The frequency of the favorable genotype (GG) for density increased 15-fold in the validation population, suggesting that breeding selection for growth and form may have influenced allele frequency in this population. Association with growth (ring width) was also detected for SNP 60 in the validation population, but the effect was negatively correlated with density.
TABLE 4.
Significant associations identified from 23 SNPs retested in the Flynn validation population
| SNP | Gene | Trait | Description | Genotype | P-value | q-value | R |
|---|---|---|---|---|---|---|---|
| 60a | pal1 | Trait 81 | Density | c/t | 0.0010 | 0.0435 | 0.039 |
| 60a | pal1 | Trait 71 | Ring Width | c/t | 0.0035 | 0.0435 | 0.037 |
| 60a | pal1 | Trait 79 | Density | c/t | 0.0320 | 0.0653 | 0.029 |
| 60a | pal1 | Trait 65 | Density | c/t | 0.0330 | 0.0653 | 0.030 |
| 64 | pcber2 | Trait 65 | Density | g/a | 0.0045 | 0.1798 | 0.035 |
| 64 | pcber2 | Trait 8 | Density | g/a | 0.0155 | 0.1914 | 0.030 |
| 64 | pcber2 | Trait 79 | Density | g/a | 0.0245 | 0.1914 | 0.030 |
| 64 | pcber2 | Trait 81 | Density | g/a | 0.0305 | 0.1914 | 0.032 |
| 146 | susy | Trait 85 | MFA | g/t | 0.0015 | 0.1914 | 0.038 |
| 146 | susy | Trait 40 | MFA | g/t | 0.0300 | 0.1914 | 0.029 |
| 146 | susy | Trait 66 | MFA | g/t | 0.0415 | 0.2188 | 0.028 |
| 146 | susy | Trait 84 | MFA | g/t | 0.0495 | 0.2393 | 0.029 |
R, proportion of the additive genetic variance controlled by SNP.
SNP shortlisted in discovery population.
Figure 3.—
Mean and standard error for trait values categorized by genotype for two SNPs in the discovery population (left) and the validation population (right). In each case associations with density were detected in both populations. Gene action observed for SNP 60 was additive in all cases. In the case of SNP 64 the gene model was dominant in the validation population; however, the effect of the GG homozygote “flipped” between populations.
In the case of SNP 64, associations were detected in the validation population with density, which was strongly correlated (R = 0.79) with specific surface area (trait 39), the associated trait in the discovery population. Although not significant, the trend across genotypes was the same for average density (trait 8) in the discovery population (Figure 3). The allele effect was only partially conserved between populations: i.e., the effect of the AA homozygote and the AG heterozygote remained constant (conferring lower and higher trait values, respectively), while the GG homozygote conferred lower trait values in the discovery population and trait values that were closer to those of the heterozygote in the validation population. Although it remains to be demonstrated, inconsistency in allele effect seen here may reflect between-site environmental interactions. In the case of SNP 146, associations with MFA were detected in the validation population, but this trait was not associated in the discovery population and was only moderately correlated with density (trait 4).
DISCUSSION
Associations with wood quality:
In the present study, 10 SNP markers were identified in the natural populations of radiata pine that were significantly associated with solid wood phenotypes after accounting for population genetic structure and multiple testing. Of these SNPs, 3 were also associated in a second population, although only 2 of these validated associations detected in the discovery population. In these cases validation provided a mechanism to verify associations and improve confidence in the SNP effect detected. The effects attributed to SNP alleles were small, in line with association studies in other tree species (Thumma et al. 2005, 2009; Gonzalez-Martinez et al. 2007, 2008; Ingvarsson et al. 2008; Eckert et al. 2009), and consistent with their quantitative nature. The number of significant markers identified in the discovery population was high compared to that in other association studies of solid wood traits (Thumma et al. 2005, 2009; Gonzalez-Martinez et al. 2007), which may bring into question those associations where validation failed. With the exception of SNP 60, markers yielding solid wood associations did not significantly affect growth, despite strong negative phenotypic correlations between these traits (Baltunis et al. 2007). This illustrates the utility of analyzing individual genetic components underlying wood variation to break negative correlations with growth that currently limit breeding programs.
Multiple associations (SNP 60 and SNP 61) were detected with phenylalanine ammonia lyase (PAL1), a lignin biosynthetic gene, homologous to ATPAL1 (AT2G37040), and the association with SNP 60 was detected in a second population. PAL1 catalyzes the conversion of phenylalanine to cinnamic acid, and its activity influences lignin content (Nakashima et al. 1997). As a major constituent of the cell wall, lignin content is expected to influence wood density, and this is the first report of a PAL gene modulating this trait. The decrease in juvenile wood density associated with the minor allele of SNP 60 (T allele: 0.14) appears to be additive and suggests a mildly deleterious effect on gene function that may be under weak negative purifying selection in the natural populations. Comparison with nine other Pinaceae species reveals that the “T” allele is not unique to radiata pine. This allele appears in at least two subgenera of Pinaceae (Figure S6), although its absence in spruce suggests that this allele may have been derived among pines. The overall low frequency of the T allele may reflect an evolutionary response to the associated phenotype. Density may be an important indicator of the hydraulic properties of wood that are under selection and has been shown to affect conductivity and drought survival in conifers, and high density may be an important adaptive trait linked to the prevention of vessel cavitation (Martinez-Meier et al. 2008; Dalla-Salda et al. 2009). Both PAL1 polymorphisms occur in exonic sequences and are unlinked, and although SNP 60 lies within the PAL-HAL active domain, it is synonymous and does not interfere with the substrate binding sites (Schwede et al. 1999). Silent SNPs can afford change via other mechanisms; for instance, a SNP in a cis-acting regulatory element is shown to influence allelic expression of COBRA in eucalyptus (Thumma et al. 2009), and further examples are emerging from association studies of human diseases (Veyrieras et al. 2008). Application of this SNP for breeding selection may require a trade-off on growth, since the favorable allele for density was associated with a 14% drop in growth for ring 4; however, associations were not detected for lower growth in the other rings.
The association with intronic SNP 64 from phenylcoumaran benzylic ether reductase (PCBER), a homolog of isoflavone reductase (AT4G39230.1), was detected in both populations and affected wood density and specific surface area. Although not shortlisted in the discovery population, the q-value for this association was low and near significant (q ∼ 0.1). The role of PCBER in wood development is not well established, but has previously been suggested to affect lignification of woody tissues due to its association with phenylpropanoid biosynthesis in poplar (Vander Mijnsbrugge et al. 2000a,b). In P. taeda PCBER is localized to the secondary xylem parenchyma (Kwon et al. 2001), where it presumably catalyzes synthesis of protective secondary metabolites such as lignans. Downregulation of PCBER has been reported to reduce lignin content (Boerjan et al. 2006), providing a possible mechanism for modulation of wood density. Similar to PAL1, selection against lower density associated with the minor allele (A allele: 11.9) in the natural populations could reflect the importance of density to vascular fitness.
Other associations:
The strong association with Rac13 that cannot be tested in the validation population remains of interest. RAC13 is homologous to a small GTP-binding protein (RAC13) that has been associated with the onset of cellulose synthesis in secondary walls of cotton (Delmer et al. 1995; Potikha et al. 1999), Arabidopsis (ATRAC2/ROP7: AT5G45970) (Brembu et al. 2005), and Zinnia (ZeRAC2) (Nakanomyo et al. 2002) and accumulates preferentially in tracheary elements. An association between an intronic RAC13 site (SNP2) and MFA of secondary cell wall cellulose of mature wood fibers was first identified in the discovery population where the minor allele (C allele: 0.072) was linked additively with low MFA (Figure S7), controlling >6% of the trait variation. This SNP was highly structured (FST = 30.3%) and was an outlier compared to the other SNPs in this data set (Figure S4), indicating that diversifying selection may have occurred at this locus. SNP 2 occurs at low frequency in Año Nuevo (C allele: 0.006) and Monterey (C allele: 0.002), but was common in Cambria (C allele: 0.38), and it is not surprising that this SNP does not segregate in the validation population, whose founders were primarily from Año Nuevo and Monterey. Importantly, SNP 2 was in low LD with the other five SNPs typed across this gene (R2 = 0.0002–0.0127), suggesting that selection may have acted locally on the intron where SNP 2 resides. When examined within Cambria, in isolation from structure (FST = 4%), both the association and the effect of the C allele were reproduced (Table 3), controlling up to 9% of variation in MFA. Low MFA is considered advantageous from a commercial forestry perspective as it results in wood with greater strength. Adaptive selection in natural populations may have similarly favored trees with improved wood strength via modulation of MFA at the Cambria site. The mechanism by which RAC13 might alter MFA is not established; however, related RAC proteins have been found to regulate cytoskeletal structure (Zhang et al. 2005), and indeed ATRAC2 has been shown to affect polar cell expansion, providing a possible path for modulation of cellular MFA (Wightman and Turner 2008).
Several of the genes harboring significant associations in this study have recently been associated with wood traits in other conifer species or show evidence of adaptive selection where fiber reinforcement may be a driving factor. Two polymorphic sites within AGP4, a cell wall arabinogalactan protein, were associated with late wood density and cell population in the mature and juvenile wood, respectively, in the discovery population, and this gene has been shown to be under diversifying selection in environmentally contrasted P. pinaster populations (Eveno et al. 2008). A single SNP from LP5 that shows signatures of strong diversifying selection in the natural populations of radiata pine (data not shown) was associated with fiber wall thickness and specific surface area in both the whole-core and mature wood fractions. An LP5-like gene was previously associated with water use efficiency in P. taeda (Gonzalez-Martinez et al. 2008). Both COMT2 and CAD, genes in the lignin biosynthetic pathway, harbored associations in the discovery population with traits including fiber wall thickness, specific surface area, early wood density and MOE, late wood MOE, and density averaged over the whole core. These genes were previously shown to influence similar traits (early and late wood density) in P. taeda (Gonzalez-Martinez et al. 2007).
Factors affecting power:
Wood quality traits are quantitative in nature and are regulated by many genes of small individual effect (Neale and Savolainen 2004). In the present study a small proportion of the total phenotypic variation for each trait type was accounted for. In part this results from incomplete sampling of the candidate gene network in combination with low genomic LD. Power to detect SNPs of small effect across multiple populations is further limited by constraints on trial design. Population sizes of many thousands have been recommended from simulation experiments to be necessary to detect associations in trees (Purcell et al. 2003; Ball 2005; Gordon and Finch 2005), and human studies employing large populations commonly detect SNPs with small effects (<2%). Phenotypic variation arising from diverse gene-by-environment interactions within and between sites was not accounted for in this study, whereas the discovery (Snowy Mountain Range) and validation (coastal valley in Southern Victoria) trials are strongly contrasted with respect to altitude, evapotranspiration, temperature, and soil type (New LocClim V 1.10). Quantitative evidence suggests that significant gene-by-environment interactions are limited for solid wood traits in radiata pine (Baltunis et al. 2007; Wielinga et al. 2009). Although overall rankings may not change for quantitative studies that examine all loci simultaneously, such interactions could affect correlation of phenotype and allele segregation for individual SNPs and might explain the “flip” in effect of the GG homozygote for SNP64. Improved power to detect and validate associations in future experiments could be achieved by (1) ensuring populations are sufficiently large, (2) designing trials such that within-site and between-site environmental interactions can be accounted for, (3) avoiding genetically structured material, and (4) establishing validation populations with families, or clonal material, from the discovery population. Tailoring region-specific suites of markers for breeding selection may be necessary where SNP effects are sensitive to local environment. Markers such as SNP 60 (PAL1) and SNP 64 (PCBER1) are robust and will have greatest utility in breeding programs.
Acknowledgments
We sincerely thank Bala Thumma for reviewing the manuscript, Colin Matheson and Charlie Bell for their insightful discussions, and Philippe Matter for technical support. We also thank the Neale laboratory at University of California, Davis, for kindly providing primers for several candidate genes. This research was funded by the Commonwealth Scientific and Industrial Research Organisation, ArborGen LLC, Forest and Wood Products Australia, and the Southern Tree Breeding Association.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.116582/DC1.
References
- Ahuja, M. R., and D. B. Neale, 2005. Evolution of genome size in conifers. Silvae Genet. 54 126–137. [Google Scholar]
- Allona, I., M. Quinn, E. Shoop, K. Swope, S. St. Cyr et al., 1998. Analysis of xylem formation in pine by cDNA sequencing. Proc. Natl. Acad. Sci. USA 95 9693–9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranzana, M. J., S. Kim, K. Y. Zhao, E. Bakker, M. Horton et al., 2005. Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet. 1 531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball, R. D., 2005. Experimental designs for reliable detection of linkage disequilibrium in unstructured random population association studies. Genetics 170 859–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball, R. D., 2007. Statistical Analysis and Experimental Design. Springer Science+Business Media, New York.
- Baltunis, B. S., H. X. Wu and M. B. Powell, 2007. Inheritance of density, microfibril angle, and modulus of elasticity in juvenile wood of Pinus radiata at two locations in Australia. Can. J. For. Res. 37 2164–2174. [Google Scholar]
- Boerjan, W., A. Polle and K. Vander Mijnsbrugge, 2006. Role in lignification and growth for plant phenylcoumaran benzylic ether reductase, U.S. patent 2006/0015967.
- Bradbury, P. J., Z. Zhang, D. E. Kroon, T. M. Casstevens, Y. Ramdoss et al., 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23 2633–2635. [DOI] [PubMed] [Google Scholar]
- Brembu, T., P. Winge and A. M. Bones, 2005. The small GTPase AtRAC2/ROP7 is specifically expressed during late stages of xylem differentiation in Arabidopsis. J. Exp. Bot. 56 2465–2476. [DOI] [PubMed] [Google Scholar]
- Burton, P. R., D. G. Clayton, L. R. Cardon, N. Craddock, P. Deloukas et al., 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cato, S., L. McMillan, L. Donaldson, T. Richardson, C. Echt et al., 2006. Wood formation from the base to the crown in Pinus radiata: gradients of tracheid wall thickness, wood density, radial growth rate and gene expression. Plant Mol. Biol. 60 565–581. [DOI] [PubMed] [Google Scholar]
- Dalla-Salda, G., A. Martinez-Meier, H. Cochard and P. Rozenberg, 2009. Variation of wood density and hydraulic properties of Douglas-fir (Pseudotsuga menziesii (Mirb.) Franco) clones related to a heat and drought wave in France. For. Ecol. Manage. 257 182–189. [Google Scholar]
- Delmer, D. P., J. R. Pear, A. Andrawis and D. M. Stalker, 1995. Genes encoding small Gtp-binding proteins analogous to mammalian rac are preferentially expressed in developing cotton fibers. Mol. Gen. Genet. 248 43–51. [DOI] [PubMed] [Google Scholar]
- Devey, M. E., S. D. Carson, M. F. Nolan, A. C. Matheson, C. T. Riini et al., 2004. a QTL associations for density and diameter in Pinus radiata and the potential for marker-aided selection. Theor. Appl. Genet. 108 516–524. [DOI] [PubMed] [Google Scholar]
- Devey, M. E., K. A. Groom, M. F. Nolan, J. C. Bell, M. J. Dudzinski et al., 2004. b Detection and verification of quantitative trait loci for resistance to Dothistroma needle blight in Pinus radiata. Theor. Appl. Genet. 108 1056–1063. [DOI] [PubMed] [Google Scholar]
- Doyle, J. J., and J. L. Doyle, 1990. Isolation of plant DNA from fresh tissue. Focus 12 13–15. [Google Scholar]
- Dungey, H. S., A. C. Matheson, D. Kain and R. Evans, 2006. Genetics of wood stiffness and its component traits in Pinus radiata. Can. J. For. Res. 36 1165–1178. [Google Scholar]
- Eckert, A. J., A. D. Bower, J. L. Wegrzyn, B. Pande, K. D. Jermstad et al., 2009. Association genetics of coastal Douglas Fir (Pseudotsuga menziesii var. menziesii, Pinaceae). I. Cold-hardiness related traits. Genetics 182 1289–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, R., R. P. Kibblewhite and S. Stringer, 1997. Kraft pulp fibre property prediction from wood properties in eleven radiata pine clones. Appita J. 50 25–33. [Google Scholar]
- Eveno, E., C. Collada, M. A. Guevara, V. Leger, A. Soto et al., 2008. Contrasting patterns of selection at Pinus pinaster Ait. drought stress candidate genes as revealed by genetic differentiation analyses. Mol. Biol. Evol. 25 417–437. [DOI] [PubMed] [Google Scholar]
- Gapare, W. J., H. X. Wu and A. Abarquez, 2006. Genetic control of the time of transition from juvenile to mature wood in Pinus radiata D. Don. Ann. For. Sci. 63 871–878. [Google Scholar]
- Gonzalez-Martinez, S. C., N. C. Wheeler, E. Ersoz, C. D. Nelson and D. B. Neale, 2007. Association genetics in Pinus taeda L. I. Wood property traits. Genetics 175 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Martinez, S. C., D. Huber, E. Ersoz, J. M. Davis and D. B. Neale, 2008. Association genetics in Pinus taeda L. II. Carbon isotope discrimination. Heredity 101 19–26. [DOI] [PubMed] [Google Scholar]
- Gordon, D., and S. J. Finch, 2005. Factors affecting statistical power in the detection of genetic association. J. Clin. Invest. 115 1408–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy, O. J., and X. Vekemans, 2002 SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2 618–620.
- Henderson, C. R., 1975. Best linear unbiased estimation and prediction under a selection model. Biometrics 31 423–447. [PubMed] [Google Scholar]
- Ingvarsson, P. K., M. V. Garcia, V. Luquez, D. Hall and S. Jansson, 2008. Nucleotide polymorphism and phenotypic associations within and around the phytochrome B2 locus in European aspen (Populus tremula, Salicaceae). Genetics 178 2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karhu, A., C. Vogl, G. F. Moran, J. C. Bell and O. Savolainen, 2006. Analysis of microsatellite variation in Pinus radiata reveals effects of genetic drift but no recent bottlenecks. J. Evol. Biol. 19 167–175. [DOI] [PubMed] [Google Scholar]
- Kass, R. E., and A. E. Raftery, 1995. Bayes factors. J. Am. Stat. Assoc. 90 773–795. [Google Scholar]
- Keurentjes, J. J. B., M. Koornneef and D. Vreugdenhil, 2008. Quantitative genetics in the age of omics. Curr. Opin. Plant Biol. 11 123–128. [DOI] [PubMed] [Google Scholar]
- Kibblewhite, R. P., 1999. Designer fibres for improved papers through exploiting genetic variation in wood microstructure. Appita J. 52 429. [Google Scholar]
- Kumar, S., 2004. Genetic parameter estimates for wood stiffness, strength, internal checking, and resin bleeding for radiata pine. Can. J. For. Res. 34 2601–2610. [Google Scholar]
- Kwon, M., L. B. Davin and N. G. Lewis, 2001. In situ hybridization and immunolocalization of lignan reductases in woody tissues: implications for heartwood formation and other forms of vascular tissue preservation. Phytochemistry 57 899–914. [DOI] [PubMed] [Google Scholar]
- Li, X., H. Wu, S. Dillon and S. G. Southerton, 2009. Generation and analysis of expressed sequence tags from six developing xylem libraries in Pinus radiata D. Don. BMC Genomics 10 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiselle, B. A., V. L. Sork, J. Nason and C. Graham, 1995. Spatial genetic-structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am. J. Bot. 82 1420–1425. [Google Scholar]
- Long, A. D., and C. H. Langley, 1999. The power of association studies to detect the contribution of candidate genetic loci to variation in complex traits. Genome Res. 9 720–731. [PMC free article] [PubMed] [Google Scholar]
- Lu, J., H. Y. Zhao, J. H. Wei, Y. K. He, C. Shi et al., 2004. Lignin reduction in transgenic poplars by expressing antisense CCoAOMT gene. Prog. Nat. Sci. 14 1060–1063. [Google Scholar]
- MacKay, J. J., D. M. Omalley, T. Presnell, F. L. Booker, M. M. Campbell et al., 1997. Inheritance, gene expression, and lignin characterization in a mutant pine deficient in cinnamyl alcohol dehydrogenase. Proc. Natl. Acad. Sci. USA 94 8255–8260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock, R., 2001. Analyse—software for processing wood quality data from Silviscan.
- Martinez-Meier, A., L. Sanchez, M. Pastorino, L. Gallo and P. Rozenberg, 2008. What is hot in tree rings? The wood density of surviving Douglas-firs to the 2003 drought and heat wave. For. Ecol. Manage. 256 837–843. [Google Scholar]
- Matheson, A. C., M. E. Devey, T. L. Gordon, W. Werner, D. R. Vogler et al., 2006. Heritability of response to inoculation by pine pitch canker of seedlings of radiata pine. Aust. For. 69 101–106. [Google Scholar]
- Nakanomyo, I., B. Kost, N. H. Chua and H. Fukuda, 2002. Preferential and asymmetrical accumulation of a Rac small GTPase mRNA in differentiating xylem cells of Zinnia elegans. Plant Cell Physiol. 43 1484–1492. [DOI] [PubMed] [Google Scholar]
- Nakashima, J., K. Takabe, M. Fujita and H. Saiki, 1997. Inhibition of phenylalanine ammonia-lyase activity causes the depression of lignin accumulation of secondary wall thickening in isolated Zinnia mesophyll cells. Protoplasma 196 99–107. [Google Scholar]
- Neale, D. B., and O. Savolainen, 2004. Association genetics of complex traits in conifers. Trends Plant Sci. 9 325–330. [DOI] [PubMed] [Google Scholar]
- Newton-Cheh, C., and J. N. Hirschhorn, 2005. Genetic association studies of complex traits: design and analysis issues. Mutat. Res. 573 54–69. [DOI] [PubMed] [Google Scholar]
- Oh, S., S. Park and K. H. Han, 2003. Transcriptional regulation of secondary growth in Arabidopsis thaliana. J. Exp. Bot. 54 2709–2722. [DOI] [PubMed] [Google Scholar]
- Olsen, K. M., S. S. Halldorsdottir, J. R. Stinchcombe, C. Weinig, J. Schmitt et al., 2004. Linkage disequilibrium mapping of Arabidopsis CRY2 flowering time alleles. Genetics 167 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oraguzie, N. C., and P. L. Wilcox, 2007. An Overview of Association Mapping. Springer Science+Business Media, New York.
- Paux, E., V. Carocha, C. Marques, A. M. de Sousa, N. Borralho et al., 2005. Transcript profiling of Eucalyptus xylem genes during tension wood formation. New Phytol. 167 89–100. [DOI] [PubMed] [Google Scholar]
- Pavy, N., C. Paule, L. Parsons, J. A. Crow, M. J. Morency et al., 2005. Generation, annotation, analysis and database integration of 16,500 white spruce EST clusters. BMC Genomics 6 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potikha, T. S., C. C. Collins, D. I. Johnson, D. P. Delmer and A. Levine, 1999. The involvement of hydrogen peroxide in the differentiation of secondary walls in cotton fibers. Plant Physiol. 119 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K., and M. Przeworski, 2001. Linkage disequilibrium in humans: models and data. Am. J. Hum. Genet. 69 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K., M. Stephens and P. Donnelly, 2000. Inference of population structure using multilocus genotype data. Genetics 155 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell, S., S. S. Cherny and P. C. Sham, 2003. Genetic power calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19 149–150. [DOI] [PubMed] [Google Scholar]
- Ritland, K., 1996. Estimators for pairwise relatedness and individual inbreeding coefficients. Genet. Res. 67 175–185. [Google Scholar]
- Schwede, T. F., J. Retey and G. E. Schulz, 1999. Crystal structure of histidine ammonia-lyase revealing a novel polypeptide modification as the catalytic electrophile. Biochemistry 38 5355–5361. [DOI] [PubMed] [Google Scholar]
- Shen, R., J. B. Fan, D. Campbell, W. H. Chang, J. Chen et al., 2005. High-throughput SNP genotyping on universal bead arrays. Mutat. Res. 573 70–82. [DOI] [PubMed] [Google Scholar]
- Spokevicius, A. V., S. G. Southerton, C. P. MacMillan, D. Qiu, S. Gan et al., 2007. Beta-tubulin affects cellulose microfibril orientation in plant secondary fibre cell walls. Plant J. 51 717–726. [DOI] [PubMed] [Google Scholar]
- Storey, J. D., and R. Tibshirani, 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry, J. M., M. M. Goodman, J. Doebley, S. Kresovich, D. Nielsen et al., 2001. Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28 286–289. [DOI] [PubMed] [Google Scholar]
- Thumma, B. R., M. R. Nolan, R. Evans and G. F. Moran, 2005. Polymorphisms in cinnamoyl CoA reductase (CCR) are associated with variation in microfibril angle in Eucalyptus spp. Genetics 171 1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumma, B. R., B. A. Matheson, D. Zhang, C. Meeske, R. Meder et al., 2009. Identification of a cis-acting regulatory polymorphism in a eucalypt cobra-like gene affecting cellulose content. Genetics 183 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, C. J., J. L. Popko, M. R. Mielke, W. J. Hu, G. K. Podila et al., 1998. Suppression of O-methyltransferase gene by homologous sense transgene in quaking aspen causes red-brown wood phenotypes. Plant Physiol. 117 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerio, L., D. Carter, J. C. Rodrigues, V. Tournier, J. Gominho et al., 2003. Down regulation of cinnamyl Alcohol Dehydrogenase, a lignification enzyme, in Eucalyptus camaldulensis. Mol. Breed. 12 157–167. [Google Scholar]
- Vander Mijnsbrugge, K., H. Beeckman, R. De Rycke, M. Van Montagu, G. Engler et al., 2000. a Phenylcoumaran benzylic ether reductase, a prominent poplar xylem protein, is strongly associated with phenylpropanoid biosynthesis in lignifying cells. Planta 211 502–509. [DOI] [PubMed] [Google Scholar]
- Vander Mijnsbrugge, K., H. Meyermans, M. Van Montagu, G. Bauw and W. Boerjan, 2000. b Wood formation in poplar: identification, characterization, and seasonal variation of xylem proteins. Planta 210 589–598. [DOI] [PubMed] [Google Scholar]
- Veyrieras, J. B., S. Kudaravalli, S. Y. Kim, E. T. Dermitzakis, Y. Gilad et al., 2008. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 4 e1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielinga, B., C. A. Raymond, R. James and A. C. Matheson, 2009. Genetic parameters and genotype by environment interactions for green and basic density and stiffness of Pinus radiata D. Don estimated using acoustics. Silvae Genet. 58 112–122. [Google Scholar]
- Wightman, R., and S. R. Turner, 2008. The roles of the cytoskeleton during cellulose deposition at the secondary cell wall. Plant J. 54 794–805. [DOI] [PubMed] [Google Scholar]
- Wu, H. X., M. B. Powell, J. L. Yang, M. Ivkovic and T. A. McRae, 2007. Efficiency of early selection for rotation-aged wood quality traits in radiata pine. Ann. For. Sci. 64 1–9. [Google Scholar]
- Yuan, Z., X. Yao, D. B. Zhang, Y. Sun and H. Huang, 2007. Genome-wide expression profiling in seedlings of the Arabidopsis mutant uro that is defective in the secondary cell wall formation. J. Integr. Plant Biol. 49 1754–1762. [Google Scholar]
- Zhang, X. G., J. Dyachok, S. Krishnakumar, L. G. Smith and D. G. Oppenheimer, 2005. IRREGULAR TRICHOME BRANCH1 in Arabidopsis encodes a plant homolog of the actin-related protein2/3 complex activator Scar/WAVE that regulates actin and microtubule organization. Plant Cell 17 2314–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, K., M. Aranzana, S. Kim, C. Lister, C. Shindo et al., 2007. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 3 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]