Abstract
One of the major hurdles for the development of gene therapy for Fanconi anemia (FA) is the increased sensitivity of FA stem cells to free radical-induced DNA damage during ex vivo culture and manipulation. To minimize this damage, we have developed a brief transduction procedure for lentivirus vector-mediated transduction of hematopoietic progenitor cells from patients with Fanconi anemia complementation group A (FANCA). The lentiviral vector FancA-sW contains the phosphoglycerate kinase promoter, the FANCA cDNA, and a synthetic, safety-modified woodchuck post transcriptional regulatory element (sW). Bone marrow mononuclear cells or purified CD34+ cells from patients with FANCA were transduced in an overnight culture on recombinant fibronectin peptide CH-296, in low (5%) oxygen, with the reducing agent, N-acetyl-L-cysteine (NAC), and a combination of growth factors, granulocyte colony-stimulating factor (G-CSF), Flt3 ligand, stem cell factor (SCF), and thrombopoietin. Transduced cells plated in methylcellulose in hypoxia with NAC exhibited increased colony formation compared to 21% oxygen without NAC (P < 0.03), demonstrated increased resistance to mitomycin C compared to green fluorescent protein (GFP )-transduced controls (P < 0.007), and increased survival. Thus, combining short transduction and reducing oxidative stress may enhance the viability and engraftment of gene-corrected cells in patients with FANCA.
Keywords: gene therapy, mitomycin C, reducing agent, hypoxia
INTRODUCTION
Fanconi anemia (FA) is an autosomal recessive or rarely, X-linked syndrome, characterized by bone marrow failure, congenital anomalies, and the early development of malignancies. Nearly all patients develop aplastic anemia, and nearly half of patients develop myelodysplastic syndrome or acute myeloid leukemia. Early mortality is related to complications from bone marrow failure, including bleeding or infection.
FA cells have a defect in DNA repair that leads to increased spontaneous chromosomal breakage and a marked sensitivity to bifunctional cross-linking agents such as mitomycin C (MMC) and diepoxybutane.1 FA can be classified into at least thirteen complementation groups, including A, B, C, D1, D2, E, F, G, I, J, L, M and N.2,3 The complementation is based upon correction of the chromosomal sensitivity to cross-linking agents in hybrid cells. Most of the FA proteins form a multi-subunit complex believed to be involved in DNA repair. FANCA accounts for approximately 65–70% of FA patients.3
There have been several studies to correct the defect of hematopoietic stem cells in FA by gene transfer, both in the laboratory and in patients.4–10 It has been long recognized that FA patient cells do not survive well in ex vivo culture. This may in part be due to sensitivity to oxygen-mediated damage, as cells could be successfully grown in a low oxygen environment.11 An extensive analysis of the potential mechanisms to enhance FA cell viability in vitro demonstrated that a combination of low oxygen and use of the reducing agent, NAC, but not amifostine [2-[(3-aminopropyl)amino]ethanethiol dihydrogen phosphate] or Vastarel [(2,3,4-trimethoxybenzyl)piperazin dihydrochloride], increased growth in clonogenic assays, permitted long term growth of LTC-ICs (long-term colony-initiating cells), and permitted engraftment of FA progenitors pre-treated in these culture conditions in NOD/SCID mice.7 Moreover, fibroblasts, lymphoblastoid cells, or hematopoietic cells from FA patients were able to be transduced with a bicistronic retroviral vector containing the cDNA for FANCA, phenotypically corrected and able to survive MMC exposure.7
The most recent gene therapy clinical trial in FANCA patients utilized retrovirus-mediated gene transfer.8 Unfortunately, successful, durable engraftment of gene-modified cells was not achieved. The attempts were also thwarted by inability to adequately mobilize hematopoietic stem cells in these patients with bone marrow failure. Critical to the future success of gene therapy in FA patients is the development of a rapid transduction protocol that will minimize the time in culture for fragile FA stem cells, achieve high transduction efficiency, ensure therapeutic expression levels of the transgene, and preserve the engraftment capability. Lentiviral vectors provide an advantage in that there can be shorter transduction time, minimizing ex vivo culture, and high level transgene expression in hematopoietic cells. A study of lentiviral transduction of murine Fanca−/− hematopoietic stem cells demonstrated the ability to successfully transduce with an MOI of 100, with preservation of engraftment capability and phenotypic correction.9 Another group reported successful in vitro correction of FA patient cells with lentiviral transduction and improved recovery of CD34+ cells using hydroxyethyl starch isolation of white blood cells.10 Thus, lentiviral transduction appears to be a promising advance.
Certainly, another critical obstacle to be overcome is the potential for insertional mutagenesis, and various strategies for minimizing the risk of vector-mediated dysregulation of nearby genes are being evaluated to reduce cancer risk. Lentiviral vectors do not integrate close to the promoters of transcriptionally active genes as frequently as gammaretroviral vectors, which is another potential advantage of lentiviral vectors.12,13
Our goal was to develop methodology that could be directly applied in a gene therapy clinical trial. Here, we have combined an abbreviated procedure for lentiviral transduction of CD34+ cells, with ex vivo cell culture in hypoxic conditions in the presence of the reducing agent NAC to improve viability and engraftment potential of human cells, for the purpose of demonstrating the feasibility and effectiveness of this approach in viral-mediated gene therapy for FA patients.
RESULTS
Transduction of normal G-CSF mobilized peripheral blood mononuclear cells, lineage depleted mononuclear cells, and CD34+ cells
We transduced cells derived from normal mobilized peripheral blood progenitor cells using a lentiviral GFP vector with an MOI of 10. Using a custom mixture of antibodies to CD3, CD14, CD19 and CD56, we removed cells that expressed these antigens, and our average yield was 16% of the initial starting mononuclear cell population after density depletion. Our average yield of CD34+ cells was 1.6% of the starting mononuclear cell population, and the average purity was 84.4% by flow cytometry. The transduction efficiency by flow cytometry for GFP positive cells was 8.7% for mononuclear cells, 6.4% for lineage depleted cells, and 5.5% for CD34+ cells. However, the transduction efficiency in colony forming cells as assessed by GFP expression was 12.5% for mononuclear cells,16% for lineage depleted cells, and 27.7% for CD34+ cells. The transduction efficiency in colony forming cells as assessed by polymerase chain reaction (PCR) was 12.5% for mononuclear cells, 18.8% for lineage depleted cells, and 10.4% for CD34+ cells. These values for transduction efficiency all represent mean values for the different samples of normal, G-CSF mobilized peripheral blood progenitor cells.
Correction of defective FANCA lymphoblasts using FancA-sW
Human lymphoblasts derived from a FANCA patient were transduced overnight with a FANCA or a GFP-expressing lentiviral vector at a multiplicity of infection (MOI) of 10. The transduced cells were subsequently exposed to increasing doses of MMC and viable cell numbers were obtained 2 and 4 days after initial exposure to MMC (Figure 2). At Day 2, cell number differences between the lymphoblasts transduced with FancA-sW versus those transduced with the GFP vector were statistically different in all concentrations of MMC. The p values were 0.0015, 0.0034, 0.0091, 0.022 for 5, 10, 20, and 50 nM of MMC, respectively. Likewise, Day 4 also had a statistically different number of viable cells in all MMC concentrations, with higher statistical differences than that observed at Day 2. The p values for Day 4 were 0.0017, 7.8 × 10−5, 1.90 × −5, and .0019 for 5, 10, 20, and 50 nM of MMC, respectively. Without MMC, there was no difference in cell viability between lymphoblasts transduced with the two vectors (p value of 1.00 for both Day 2 and Day 4), suggesting that there was no innate in vitro increase of cell growth or viability in cells transduced with the FancA-sW vector. These studies demonstrated that FancA-sW transduction can correct the hypersensitivity of human FANCA-deficient cells to MMC. These results further suggest that the housekeeping constitutive internal promoter, the PGK promoter, is sufficient to achieve phenotypic correction.
Figure 2. Transduction of defective FANCA lymphoblasts by FancA-sW restores resistance to DNA cross-linking.
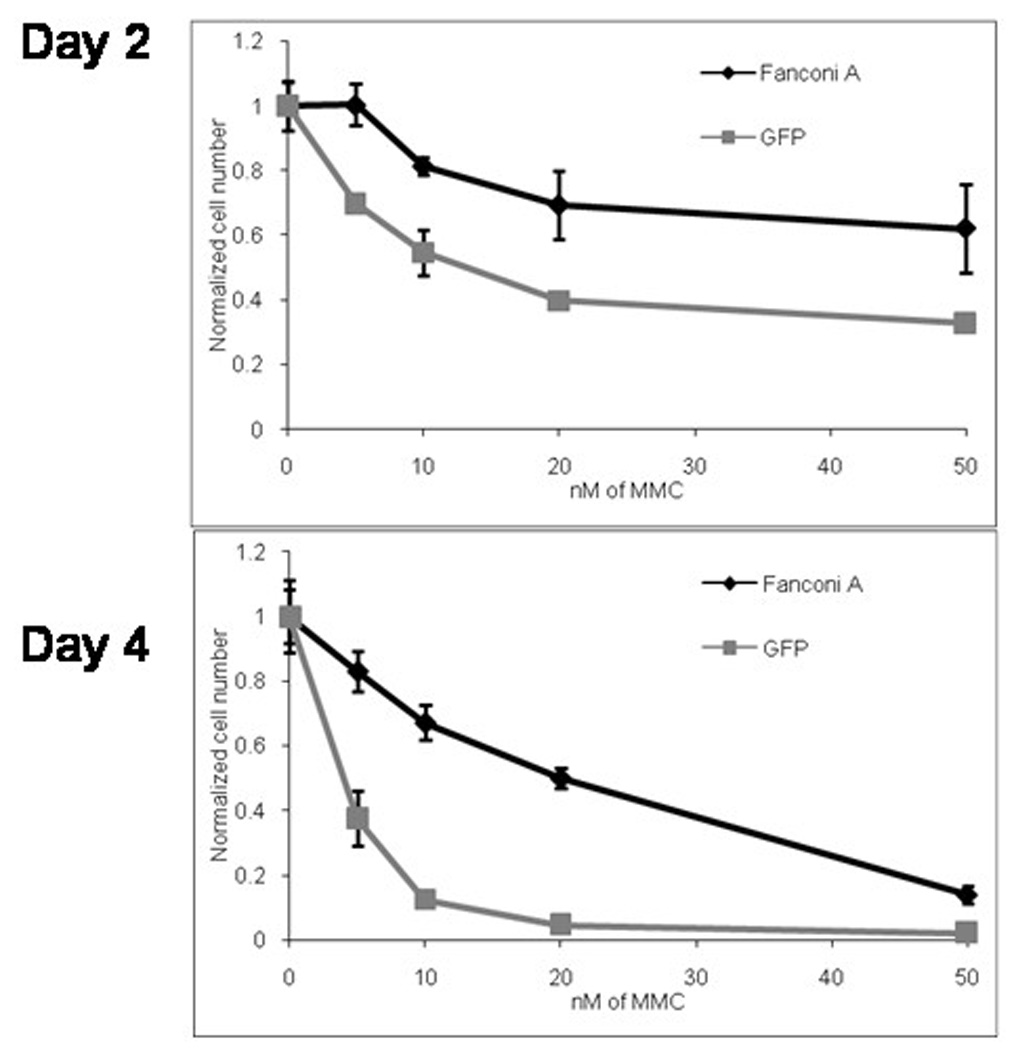
EBV-immortalized human FANCA lymphoblasts were transduced overnight with FancA-sW or control GFP vector at vector to cell ratio of 10, expanded for 2 days and then challenged with different concentrations of MMC in culture over 4 days. Cell counts were performed on days 2 and 4 of MMC culture and were normalized to untreated controls. Normalized cell number refers to ratio of viable cells at the given time in culture, compared to the starting cell number. Note that there was superior survival for the FancA-sW transduced lymphoblasts (black line) compared to the control GFP vector transduced cells (gray line).
Hypoxic environment and NAC improve transduction efficiency and viability of human FANCA CFU
One of the major technical difficulties for development of gene therapy for FA is the extraordinary ex vivo fragility of FA bone marrow cells, which is believed to be in part due to oxidative damage. We therefore sought to compare the effect of oxidative stress and length of transduction on the transduction rate and colony forming potential of primary human FA bone marrow. In order to minimize ex vivo manipulation and multiple integration events, bone marrow mononuclear cells were transduced at an MOI of 10. In an initial experiment, the effects of low oxygen, short overnight vector exposure (without pre-stimulation) and the use of NAC were compared to the standard pre-stimulation of 24 hours, followed by 2 daily cycles of transduction in normal oxygen without NAC (data not shown). Bone marrow cells transduced overnight in the presence of both low oxygen and NAC exhibited increased colony number/plate compared to conditions of 21% oxygen without NAC, mean 6.5 vs. 2 colonies per 3 × 104 cells plated (p = 0.12). Short transduction period, use of 5% oxygen and use of NAC were each associated with increased colony number, with p values of 0.15, 0.15, and 0.089, respectively. Using either short transduction or NAC was associated with a trend in having increased GFP-positive colonies (p = 0.10 for both). The use of 5% oxygen showed a significant difference compared to using 21% oxygen, with an average of 2.125 GFP colonies compared to 0.75 colonies (p = 0.018). Lastly, there was a significant difference in the number of GFP vector transduced colonies when the brief transduction, low oxygen with reducing agent was compared to long transduction, 21% oxygen and no NAC, with an average of 4 colonies/plate in the overnight low oxygen with NAC versus 0 colonies/plate in the standard conditions (p = 0.00042).These data demonstrate that short transduction with reduced oxidative stress is more effective in transducing progenitor cells and at least as effective in preserving colony forming cells compared to standard transduction conditions.
In a second experiment, we focused on the role of oxygen and NAC on transduction and colony formation in a single overnight transduction (Figure 3). The combined use of 1 mM NAC and 5% oxygen showed a significant increase in the number of colonies formed compared to the other conditions (p values all < 0.03). The greatest difference was between the colonies formed in 5% oxygen with NAC and those formed under 21% oxygen without NAC, with an average of 11.5 ± 0.71 (SD) colonies and 3 ± 0.05 (SD) colonies respectively (p = 0.0034). No other group was significantly different from another. Transduction rate, measured by GFP expression was statistically similar in the four groups (p > 0.22); with 60% of colonies placed in hypoxic conditions with NAC being GFP-positive. Combining data from the various groups, the use of NAC was associated with both increased colony formation, 8.25 versus 3 (p value of 0.038), and increase in the number of transduced colonies, 4.25 versus 0.75 (p value of 0.022). Thus the overnight (14-hour) transduction of whole bone marrow, in an environment of reduced oxygen stress, results in a high level of transduction with preservation of progenitor cells.
Figure 3. Hypoxic environment and/or reducing agent improve the transduction efficiency and viability of human Fanconi bone marrow progenitors.
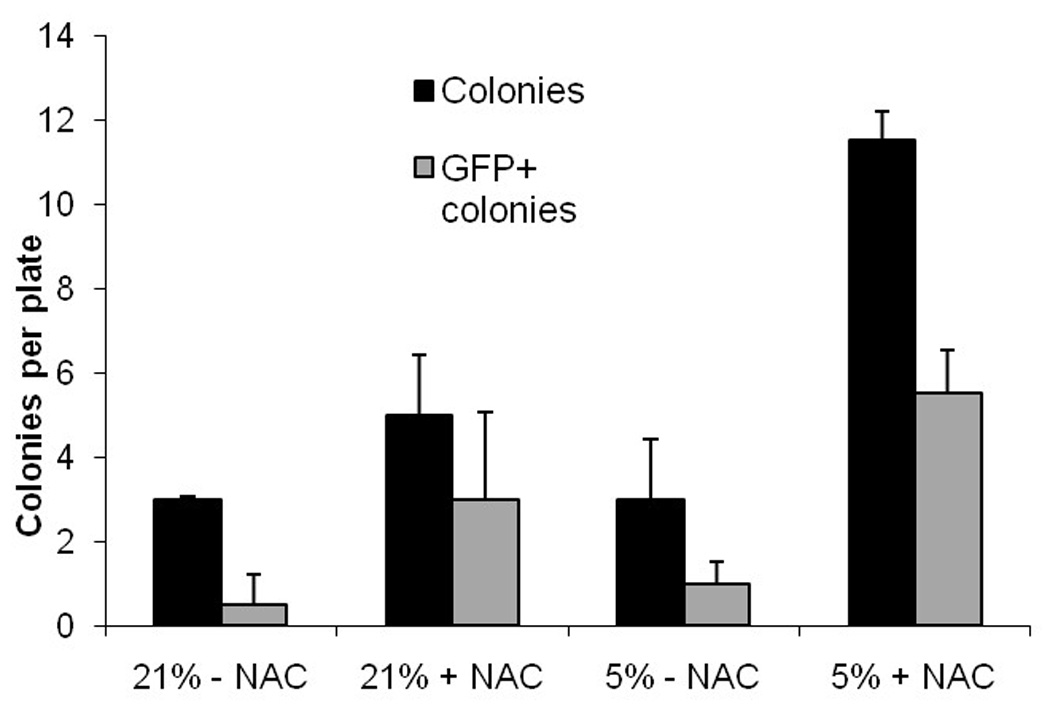
Human Fanconi bone marrow mononuclear cells were transduced overnight with the GFP vector at a vector to cell ratio of 10:1 and plated in colony assays at 20,000 cells per plate. During the transduction and colony assays, cells were exposed to either 21% or 5% O2 with or without 1mM NAC, a reducing agent. Transduction efficiency increased from 16.7% to 47.8% by reducing oxygen from 21% to 5% and adding 1 mM NAC.
Correction of human FA-A bone marrow cell MMC hypersensitivity after FancA-sW transduction
Three human FANCA bone marrow samples were used to analyze efficacy of the FancA-sW lentiviral vector. The mononuclear cells derived from these samples were transduced in 5% O2 with 1mM NAC. The cells were subsequently divided and either plated in methylcellulose or placed into suspension culture in the hypoxia chamber, with or without MMC.
While there is generally a reduced clonogenic capacity of FA hematopoietic progenitors in vitro,18–20 CFU were readily detectable in bone marrow mononuclear cells from three different FANCA patients transduced with FancA-sW as compared to GFP (mean 10.7, 54 and 14 colonies/3 × 104 cells plated versus 6, 27, and 5 colonies/3 × 104 cells plated, respectively). The distribution of colony size is shown in Figure 4A. Colony morphology was heterogeneous, including colonies greater than 2 mm and clusters of < 50 cells. On average, GFP vector transduced cells produced 52% less colonies and clusters compared to FancA-sW transduced cells (p = 0.019). While number of colonies diminished with increasing MMC concentration, there were statistically more colonies for the FancA-sW transduced cells than the GFP vector transduced cells (Figure 4B). The fraction of CD34+ cells in the patient bone marrow ranged from 0.1 to 0.7%, and did not correlate with colony formation or transduction efficiency. Transduction efficiency ranged from 15 to 90% for both FancA-SW and GFP vector transduced bone marrow by colony PCR, demonstrating effective transduction using a single, overnight transduction at an MOI of 10.
Figure 4. There are more colonies surviving in MMC for FancA-sW transduced human FANCA bone marrow cells than for the control GFP vector transduced cells.
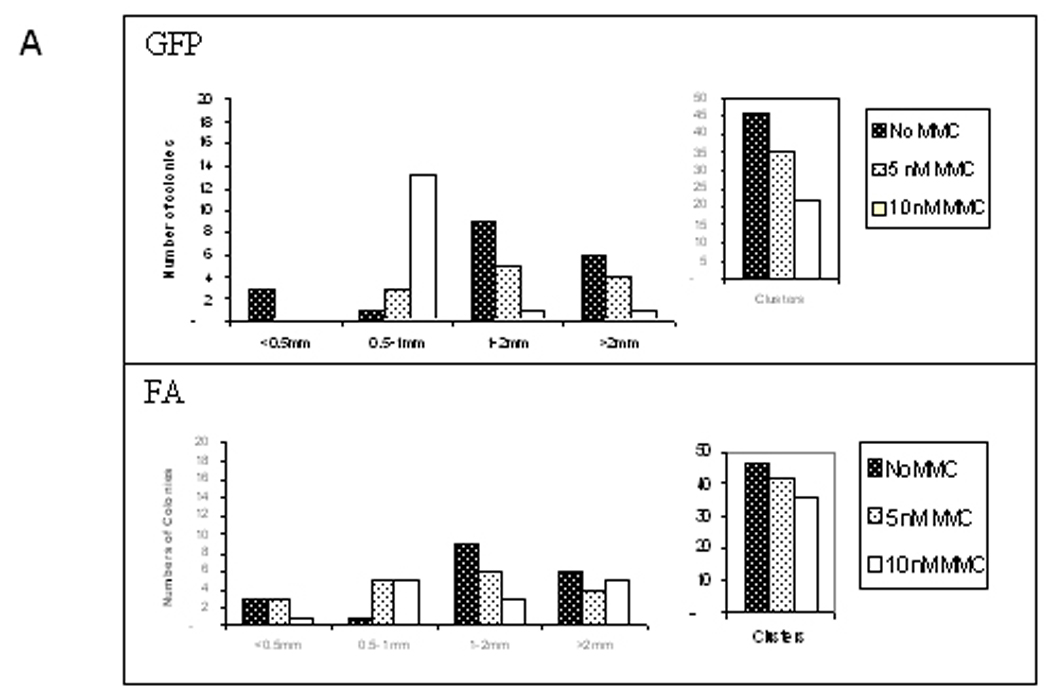
A) Patient 5 colony size distribution in increasing concentrations of MMCNote that there were fewer larger colonies in 10 nM MMC for the control GFP vector transduced cells than for the FancA-sW transduced cells. With increasing MMC concentration, there were fewer colonies or cell clusters, and the effect was more pronounced for the control GFP vector transduced cells than for the FancA-sW transduced cells. B) Patients 5, 6, and 7 colony numbers in different concentrations of MMC. There were fewer colonies for the GFP vector transduced cells than for the FancA-sW transduced cells, and the differences in colony numbers were statistically significant for the indicated MMC concentrations. For patient 5 at 0 nM MMC, p=0.035, at 10 nM MMC p=0,030, and at 20 nM p=0.26; for patient 6 at 0 nM MMC p=0.001, at 10 nM MMC p=0.009, and at 20 nM MMC p=0.45; for patient 7 at 0 nM MMC p=0.086 and at 20 nM MMC the colony numbers were too low for statistical analysis.
Transduced bone marrow cells placed in cell culture were exposed to increasing concentrations of MMC in triplicate wells. Viable cells were subsequently counted at Day 2 and Day 4 with the results shown in Figure 5. All three samples demonstrated statistically significant increased cell number in the FancA-sW transduced cells, compared to GFP vector transduced controls, at the highest level tested on both Day 2 and Day 4 (Table 1). Patient 1 showed significant differences at both 5 and 10 nM of MMC with a 2.1-fold difference in cell number in cells exposed to 10 nM MMC. At Day 4, patient 1 showed significant difference in cell number only in cells exposed to 10 nM MMC. For patient 2, Day 2 samples showed significant differences in cell numbers in all three MMC concentrations tested. On Day 4, patient 2 had significant differences in transduced cells exposed to MMC at both 10 and 20 nM concentrations. Patient 3 had GFP vector transduced cells that were the most resistant of the three samples to the effects of MMC. Overall, these results indicate that human FA bone marrow cell hypersensitivity to MMC-induced DNA cross linking is corrected by transduction of the cells with the FancA-SW vector in hypoxia with reducing agent, although the effect varied between individual patients.
Figure 5. There is superior survival of FancA-sW transduced FANCA bone marrow cells in culture in MMC than control GFP vector transduced cells.
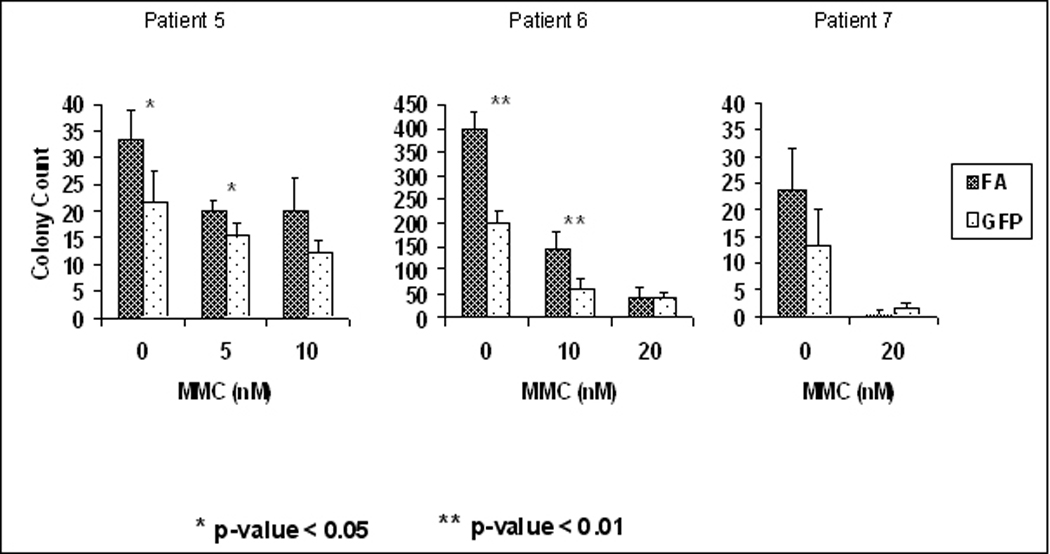
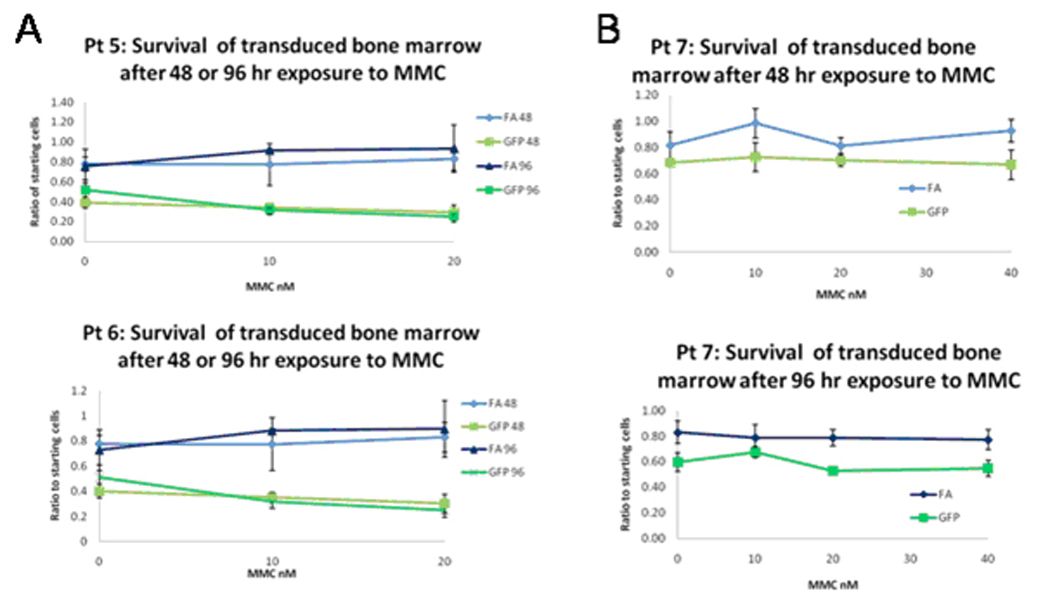
Human Fanconi bone marrow mononuclear cells were transduced overnight with FancA-sW (FA) or GFP (GFP=RRL.sin.cPPT.PGK.GFP.wpre) vector at a vector to cell ratio of 10:1. The cells were subsequently plated into 96-well plates and exposed to increasing MMC concentrations in triplicate. Viable cells were counted at 48 and 96 hours after exposure to MMC. During the transduction and MMC challenge, cells were placed in 5% O2 with 1 mM NAC. A. Survival cultures for patient 5 (upper) and patient 6 (lower) for transduced cells with the indicated vector at the indicated time of culture, 48 or 96 hours. B. Survival cultures for patient 7, upper curves at the 48 hr time point and lower curves at the 96 hr time point for the indicated vector, as above. Significant differences were determined by student t-test comparing cell survival of FancA-sW to GFP vector transduced bone marrow for the same MMC concentration at the same time point, and the p values are listed in Table 1. Blue lines indicate FancA-sW transduced cells, green lines control GFP vector transduced cells.
Table 1.
p values for differences in numbers of surviving transduced cells for FANCA-sW versus GFP vector with increasing MMC concentration
| Mitomycin C (nM) |
|||||
|---|---|---|---|---|---|
| Patient | 0 | 5 | 10 | 20 | 40 |
| Pt. 5, 2 days | 0.32 | 0.020 | 0.00034 | ||
| Pt. 5, 4 days | 0.16 | 0.11 | 0.0123 | ||
| Pt. 6, 2 days | 0.0059 | 0.047 | 0.0056 | ||
| Pt. 6, 4 days | 0.38 | 0.0002 | 0.012 | ||
| Pt. 7, 2 days | 0.087 | 0.046 | 0.0651 | 0.034 | |
| Pt. 7, 4 days | 0.024 | 0.16 | 0.0031 | 0.019 | |
Significant p values are in bold text.
Isolation and transduction of Fanconi bone marrow derived CD34+ cells
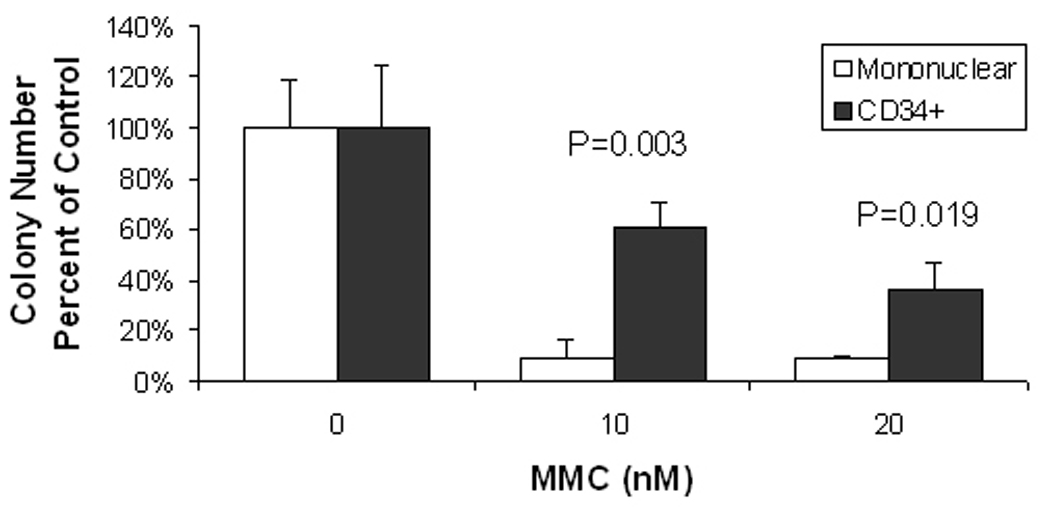
From 1.2 ml of patient bone marrow, we obtained 18 million mononuclear cells after density depletion. From a fraction of 11 million bone marrow mononuclear cells, we obtained 110,000 CD34+ cells after positive selection on the Miltenyi MACS, a yield of 1% of the mononuclear cells. Fifty thousand cells were transduced with the GFP lentiviral vector, and the same number with FancA-sW. After overnight transduction in the hypoxia chamber with NAC, 31,000 (62%) were recovered from the GFP vector transduction, and 35,000 (70%) from the FancA-sW transduction. To extrapolate the yield for a scaled up clinical procedure, for a 500 ml bone marrow harvest, this would correspond to 52,500,420 transduced CD34+ cells, or 1 million transduced bone marrow CD34+ cells/kg for a 50 kg patient, 1.3 million/kg for a 40 kg patient, or 1.7 million/kg for a 30 kg patient. The transduced CD34+ cells exhibited colony growth in methylcellulose, with survival in MMC that was superior to that of transduced mononuclear cells (Figure 6). For 10 nM MMC, there were 60.7% ± 10.1% of the colonies at 0 nM MMC for the CD34+ cells, and 8.9 ± 7.4% for mononuclear cells (p=0.003), and at 20 nM MMC, there were 35.7% ± 10.3% of the colonies at 0 nM MMC for the CD34+ cells, and 8.9% ± 1.3% for the mononuclear cells (p=0.019), suggesting that there was even better functional correction of the colony forming cells from transduced purified FA patient derived CD34+ cells than the transduced mononuclear cells. By colony PCR, 12.5% of the colonies plated from the FancA-sW transduced CD34+ cells were positive for the transgene without MMC, and 18.8% positive with 10 nM MMC.
Figure 6. Transduced CD34+ FA patient bone marrow cells form colonies and survive mitomycin C (MMC).
The x-axis indicates the MMC concentration, 0, 10 or 20 nM for the cells plated in methylcellulose. The y-axis indicates the colony number as percent of the no MMC number. Note that there are a statistically significant higher number of colonies at 10 and 20 nM MMC for the FancA-sW transduced CD34+ cells compared to transduced mononuclear cells. The p values are indicated on the graph.
Analysis of integration sites by linear amplification-mediated polymerase chain reaction (LAM-PCR)
LAM-PCR was used to analyze insertion sites for FancA-sW transduced normal mobilized progenitor cells and FANCA bone marrow cells. Insertion sites for the normal mobilized progenitor cells are exhibited in Table 2. LAM-PCR analysis of DNA from colonies derived from FancA-sW transduced human FANCA bone marrow cells demonstrated 1–2 integrations per CFU, with the majority exhibiting one integration per cell (data not shown). By BLAST analysis, the integration sites were not within coding sequences of the genome.
Table 2.
LAM-PCR Insertion site analysis: 26 insertions in bulk culture of transduced normal CD34+ cells
| Chromosome | Location | Number |
|---|---|---|
| Chromosome 1 | 70390918–70390946 | 8 |
| Chromosome 19 | 46739495–46739521 | 1 |
| Chromosome 19 | 12363991–12364068 | 1 |
| Chromosome 19 | 47409927–47409978 | 7 |
| Chromosome 3 | 52285411–52285698 | 1 |
| Chromosome 4 | 78944104–78944152 | 1 |
| Chromosome 17 | 4079092–4079145 | 2 |
| Chromosome 17 | 34879822–34879863 | 2 |
| Chromosome 6 | 3035030–3035056 | 3 |
DISCUSSION
We have developed an effective and clinically relevant method to transduce Fanconi anemia patient bone marrow cells using a safety modified lentiviral vector and a brief transduction period in a hypoxic environment with a reducing agent. This combination of conditions was able to preserve the ability to form colonies in vitro, with consistent correction of sensitivity to MMC induced double strand DNA cross-linking indicating phenotypic correction.
There are several safety features engineered into the FancA-sW vector construct relevant to its use for clinical applications. Previous FA gene therapy studies have either used the viral long terminal repeat or strong internal promoters, which could alter the expression of neighboring genes after viral integration. In order to minimize the risk for promoter enhancer effects in our construct, the FancA gene is driven by a housekeeping constitutive internal promoter, the promoter for human PGK, and a WPRE was included to increase transgene expression.14 For safety concerns, a synthetic WPRE was created which interrupted the partial open reading frame for Protein X which is present in the WPRE.15 These efforts were undertaken in order to eliminate strong viral promoters which may potently enhance the expression from nearby genes thus minimizing the risk of insertional mutagenesis leading to the activation of proto-oncogenes.
There have been several hypotheses proposed for the exact mechanism by which hypoxia improves the survival of hematopoietic progenitors in Fanconi anemia. Hematopoietic stem cells are believed to be located in a hypoxic environment within the bone marrow, in the endosteal niche,16 and there was enhanced maintenance of human cord blood stem cells capable of repopulating NOD/SCID/IL-2Rγnull immunodeficient mice after incubation in a hypoxic environment.17 In fact, hypoxia may promote stem cell survival for other tissues as well, and hypoxia-inducible factor-1alpha may play a role through its effects on cell cycle and transcription. In the case of Fanconi anemia bone marrow progenitors, reduction of oxygen to 5% was demonstrated to improve erythroid colony formation,18 and there was greatly enhanced colony formation observed for the combination of hypoxia and reducing agent.7 Another group also presented data on lentiviral transduction and colony assays in 3% oxygen for two FA patients, with variable results on transduction efficiency and improved colony growth in at least one of the patients,10 but error bars and statistical analysis were not provided.
We also found that both hypoxia and N-acetyl cysteine increased the colony forming ability of human Fanconi A hematopoietic progenitors, and that there was also improved transduction efficiency. However, MMC requires oxygen radicals for DNA cross-linking which is likely inhibited by the reduced oxygen environment of the culturing conditions resulting in the need for in higher doses of drug than was anticipated. Nevertheless, our in vitro culture assay of transduced bone marrow cells in MMC offers a rapid and relatively simple method to determine FA patient subtype (with appropriate vectors) without the need for colony assay, T cell expansion, or lymphocyte transformation.
Gene transfer to a fraction of hematopoietic progenitors may result in expansion of those clones of cells due to survival advantage with acquisition of DNA repair ability. In fact, there have been cases of spontaneous reversion in a lymphohematopoietic stem cell in Fanconi anemia, and the reverted cells dominated the marrow due to survival advantage while the patients retained mosaicism, usually with the Fanconi mutation preserved without compensatory correction in skin fibroblasts.19–22
Our studies focused on procedures for lentiviral vector transduction of Fanconi anemia progenitor cells. As it has been demonstrated that there is defective homing of FANCA bone marrow progenitors leading to deficient engraftment in NOD/scid mice due to decreased activity of the Rho GTPase cdc42,23 it will be interesting to study whether hypoxia and/or reducing agent NAC can improve this ability, as hypoxia had improved engraftment of cord blood progenitors in NOD/scid mice.17 It is of utmost importance to successful gene therapy that engraftment capability be preserved after ex vivo transduction.
These data suggest that lentivirus-mediated gene transfer in a rapid transduction protocol with hypoxia and reducing agent may sufficiently preserve hematopoietic progenitor potential so that sufficient engraftment and in vivo transgene expression may be attained in a clinical trial.
METHODS AND METHODS
FANCA lentiviral vector
A third generation self-inactivating lentiviral vector containing the full-length FANCA gene was assembled that would be suitable for a gene therapy clinical trial, designated FancA-sW (Figure 1). The lentiviral vector containing the FANCA transgene was constructed using the HIV-1–based RRL.sin.cPPT.PGK.GFP.wpre self-inactivating vector,24 which contains a human phosphoglycerate kinase promoter (PGK) driving expression of enhanced green fluorescent protein (GFP). The vector also contains a central polypurine tract (cPPT) to enhance transduction.25 Transcription is driven by a heterologous RSV promoter, and there is a 400 bp deletion of the U3 region of the 3' LTR that contains all of the major determinants responsible for regulating the HIV-1 LTR promoter including the TATA box.
Figure 1. FancA-sW lentiviral vector.

The self inactivating lentiviral vector, FancA-sW, includes a phosphoglycerate kinase (PGK) promoter, the human FancA coding sequence, and a synthetic woodchuck hepatitis virus posttranscriptional regulatory element (sW). As an additional safety feature the sW was modified to interrupt a partial woodchuck hepatitis virus X protein open reading frame. LTR is long terminal repeat, CAR is lentiviral cis-acting region.
The FANCA cDNA was kindly provided by Dr. Hans Joenje in the mammalian expression vector pREP4 (Invitrogen, Carlsbad, CA). A 5.5 kb NarI to HindIII fragment including the FANCA cDNA was transferred to the MND-EGFP SN oncoretroviral plasmid to create MNDFANCA. An AgeI site was created upstream of the FANCA open reading frame (ORF) in MNDFANCA by converting an SfoI site to an AgeI site using an AgeI linker. The 3.2 kb AgeI to BamHI fragment and 1.6 kb BamHI to BsrGI fragments, spanning 4.8 kb of the FANCA cDNA, including the 4.4 kb FANCA ORF, were cloned into the AgeI to BsrGI site of RRLsin.cPPT.hPGK.GFP.wpre replacing the GFP ORF to generate pRRL.sin.cPPT.PGK.FANCA.wpre. Finally, the wpre element was replaced with a synthetic woodchuck element (sW), interrupting the partial Protein X ORF present within the wpre sequence.
Lentiviral vector stocks of VSV-G–pseudotype lentiviral vectors were prepared transiently by calcium phosphate-mediated four-plasmid transfection of 293T cells. Briefly, 27 µg of the transfer vector construct, 17.5 µg second-generation gag-pol packaging construct pCMVΔR8.74, and 9.5 µg VSV-G expression construct pMD.G were used for transfection of 12 × 106 293T cells overnight in 25 mL Dulbecco’s Modified Eagle Medium (DMEM) with 10% heat-inactivated fetal bovine serum. The cells were treated with 10 mM sodium butyrate during the first of three 12-hour vector supernatant collections. The supernatant was filtered through a 0.22 µm-pore-size filter and concentrated 100-fold by centrifugation.
The same vector backbone containing the green fluorescent protein (GFP) cDNA instead of the FANCA cDNA, RRLsin.cPPT.hPGK.GFP.wpre (GFP vector), was used as a transduction control.
Lymphoblast transduction and MMC challenge
The human FANCA cell line HSC7226 was used to demonstrate functional efficacy of the lentiviral vector. HSC72 cells were placed on plates coated with fibronectin peptide CH-296, RetroNectin® (Takara Bio USA; Madison, WI) at 2 mcg/cm2 and incubated with virus supernatant at a multiplicity of infection (MOI) of 10 in a humidified incubator for 14 hours at 37°C, and 5% CO2.
For the culture of lymphoblasts in MMC, 30,000 cells/well were placed into 96-well plates in triplicate containing transduction media and increasing concentrations of MMC (Ben Venue Laboratories, Inc., Bedford, OH), 0–40 nM. The cells were placed in a humidified incubator with 5% CO2 at 37°. Surviving cells were enumerated by Trypan blue (MP Biomedicals, LLC: Solon, OH) exclusion 2 and 4 days later.
Isolation of normal bone marrow mononuclear cells, lineage depleted cells, and CD34+ selected cells
Mononuclear cells were isolated from G-CSF-mobilized peripheral blood of normal donor subjects by using Ficoll-Paque Plus (GE-Health Care Bio-sciences AB, Uppsala, Sweden). A proportion of the cells was then subjected to stem and progenitor cell enrichment by lineage depletion using EasySep® Human Hematopoietic Progenitor Cell Enrichment Kit (StemCell Technologies, Vancouver, BC, Canada). A custom mixture of biotinylated antibodies to CD14, CD19, CD3 and CD56 (StemCell Technologies) was utilized for depletion of cells bearing these markers in the magnetic device, and the cells lacking these markers were retained for transduction. Another portion of the collected mononuclear cells was subjected to positive selection of CD34 expressing progenitors, using the Direct CD34 Progenitor Cell Isolation MicroBead Kit (Miltenyi Biotec, Auburn, CA). The magnetic beads conjugated with mouse anti-human CD34 antibody were mixed with the mononuclear cells and incubated at 4°C for 30 minutes. The cells were then passed through MACS® Separation Column (Miltenyi Biotec) positioned in the magnetic MACS Separator to allow the unlabeled cells run through while the magnetically labeled cells are retained on the column. The column was washed twice, then removed from the Separator and the CD34+ cells were eluted. CD34 cell enrichment efficiency was verified by flow cytometry.
Isolation of bone marrow mononuclear cells from FA-A patients
Primary human bone marrow was obtained from FA patients for research purposes under protocols approved by the Institutional Review Boards at each of the participating clinical sites and the Fred Hutchinson Cancer Research Center. Subjects provided written informed consent.
Bone marrow mononuclear cells were isolated by density depletion on lymphocyte separation media as described above for normal mobilized progenitors, and CD34+ cells were also isolated for one patient, as described above for normal progenitors. The cells were washed twice in PBS prior to being resuspended in transduction media. Transduction media consisted of Iscove's Modified Dulbecco's Medium (IMDM) supplemented with penicillin/streptomycin (Cellgro; Manassas, VA), 4 mM L-glutamine, HEPES, and 20% fetal bovine serum (latter three components from Hyclone; Ogden, UT), with 100 ng/ml each of recombinant human Fms-related tyrosine kinase 3 ligand (Flt3-Ligand), stem cell factor (SCF), and thrombopoietin (Tpo), and human granulocyte colony-stimulating factor (G-CSF). The human cytokines were purchased from CellGenix; Antioch, IL, with the exception of G-CSF, which was from Amgen; Thousand Oaks, CA. The media was further supplemented with 1 mM cell culture grade N-Acetyl-L-cysteine (NAC; Sigma-Aldrich; St. Louis, MO).
Human FANCA bone marrow nuclear cell transduction and colony assay
Cells were placed in bacteriologic plates coated with RetroNectin® at 2 mcg/cm2 and incubated with the lentiviral vectors at an MOI of 10, then placed into a humidified hypoxia chamber (Billups-Rothenberg; Del Mar, CA) with 10% CO2 and 5% O2 for 14 hours at 37°C. Cells were subsequently washed off the plate, resuspended and counted.
For colony assay, 25–30,000 mononuclear cells, 3500–7,500 lineage depleted cells, or 1500 CD34+ cells were placed into 1 ml of methylcellulose (MethoCult® M3434 by StemCell Technologies), 1 mM NAC, in triplicate and placed into a humidified hypoxia chamber with 10% CO2 and 5% O2 at 37°C The colonies were counted and isolated at about 18 days.
For bulk culture MMC challenge: 25,000–30,000 cells were placed into 96-well plates in triplicate containing transduction media and increasing amounts of MMC. The cells were placed in a humidified hypoxia chamber with 10% CO2 and 5% O2 at 37°C. Cell number was determined 2 and 4 days later.
Colony Forming Unit (CFU) lentivirus-specific PCR
To determine the fraction of CFUs that exhibited integrated lentiviral transgene, DNA was isolated from methylcellulose colonies, and PCR was performed using primers specific for proviral sequence. Methylcellulose colonies were picked up with the aid of a dissecting microscope, and DNA was isolated using the QIAamp DNA Mini Kit as per manufacture's protocol (Qiagen Inc.; Valencia, CA). Approximately 25 µl of DNA extract was used for PCR. PCR conditions included 42 cycles at 94°C for one minute, 65°C for 30 seconds and 72°C for one minute, followed by a 10 min. extension at 72°C, as described.27 Negative controls include no DNA (water) or DNA isolated from non-transduced cells. The percentage of positive CFUs was derived from the fraction of those with PCR products for the lentiviral proviral sequence and positive for control, glyceraldehyde phosphate dehydrogenase (GAPDH), over the total number positive for positive control GAPDH only. The following primers were used: Primers for the detection of lentiviral vector: LentiF: 5'-AGAGATGGGTGCGAGAGCGTCA, LentiR: 5'-TGCCTTGGTGGGTGCTACTCCTAA; Mouse DNA Control: Beta actin Forward: 5'-GGT GAT AAG TGG CCT TGG AG-3' Reverse: 5'-CTG AGA CAT GCA AGG AGT GC-3' Human DNA Control: GAPDH Forward: 5'-TCT AGA CGG CAG GTC AGG TCC ACC-3' Reverse: 5'-CCA CCC ATG GCA AAT TCC ATG GCA-3'.
Linear amplification (LAM)-PCR
Lentiviral integration site analysis was performed by LAM-PCR on DNA obtained from the methylcellulose colonies, or transduced mononuclear cells, as previously described.13 One hundred nanograms of DNA served as the template.
Statistical analysis
p values were determined by Student’s t test.
Figure 7.
ACKNOWLEDGMENTS
Grant Support This work was supported by NIH grants HL085693, DK56465 and DK47754 to HPK. We would like to thank Helen Crawford and Bonnie Larson for help with the preparation of the manuscript. HPK is a Molecular Medicine Investigator and the recipient of the José Carreras/ E. Donnall Thomas Endowed Chair for Cancer Research.
Grant Support: National Institutes of Health
Footnotes
Portions of this work were presented in preliminary form at the American Society of Gene Therapy and American Society of Hematology meetings, 2008.
Conflicts of Interest: None.
REFERENCES
- 1.Bagby GC, Alter BP. Fanconi anemia (Review) Semin Hematol. 2006;43:147–156. doi: 10.1053/j.seminhematol.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Taniguchi T, D'Andrea AD. Molecular pathogenesis of Fanconi anemia: recent progress (Review) Blood. 2006;107:4223–4233. doi: 10.1182/blood-2005-10-4240. [DOI] [PubMed] [Google Scholar]
- 3.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins (Review) Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 4.Liu JM, Kim S, Read EJ, Futaki M, Dokal I, Carter CS, et al. Engraftment of hematopoietic progenitor cells transduced with the Fanconi anemia group C gene (FANCC) Hum Gene Ther. 1999;10:2337–2346. doi: 10.1089/10430349950016988. [DOI] [PubMed] [Google Scholar]
- 5.Yamada K, Olsen JC, Patel M, Rao KW, Walsh CE. Functional correction of fanconi anemia group C hematopoietic cells by the use of a novel lentiviral vector. Mol Ther. 2001;3:485–490. doi: 10.1006/mthe.2001.0287. [DOI] [PubMed] [Google Scholar]
- 6.Yamada K, Ramezani A, Hawley RG, Ebell W, Arwert F, Arnold LW, et al. Phenotype correction of Fanconi anemia group A hematopoietic stem cells using lentiviral vector. Mol Ther. 2003;8:600–610. doi: 10.1016/s1525-0016(03)00223-5. [DOI] [PubMed] [Google Scholar]
- 7.Cohen-Haguenauer O, Peault B, Bauche C, Daniel MT, Casal I, Levy V, et al. In vivo repopulation ability of genetically corrected bone marrow cells from Fanconi anemia patients. Proc Natl Acad Sci USA. 2006;103:2340–2345. doi: 10.1073/pnas.0510613103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelly PF, Radtke S, von Kalle C, Balcik B, Bohn K, Mueller R, et al. Stem cell collection and gene transfer in Fanconi anemia. Mol Ther. 2007;15:211–219. doi: 10.1038/sj.mt.6300033. [DOI] [PubMed] [Google Scholar]
- 9.Müller LU, Milsom MD, Kim MO, Schambach A, Schuesler T, Williams DA. Rapid lentiviral transduction preserves the engraftment potential of Fanca(−/−) hematopoietic stem cells. Mol Ther. 2008;16:1154–1160. doi: 10.1038/mt.2008.67. [DOI] [PubMed] [Google Scholar]
- 10.Jacome A, Navarro S, Rio P, Yañez RM, González-Murillo A, Lozano ML, et al. Lentiviral-mediated genetic correction of hematopoietic and mesenchymal progenitor cells from Fanconi anemia patients. Mol Ther. 2009;17:1083–1092. doi: 10.1038/mt.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butturini A, Gale RP, Verlander PC, Aldler-Bricner B, Gillio AP, Auerbach AD. Hematologic abnormalities in Fanconi anemia: An International Fanconi Anemia Registry Study. Blood. 1994;84:1650–1655. [PubMed] [Google Scholar]
- 12.Laufs S, Guenechea G, Gonzalez-Murillo A, Zsuzsanna Nagy K, Luz Lozano M, del Val C, et al. Lentiviralvector integration sites in human NOD/SCID repopulating cells. J Gene Med. 2006;8:1197–1207. doi: 10.1002/jgm.958. [DOI] [PubMed] [Google Scholar]
- 13.Beard BC, Keyser KA, Trobridge GD, Peterson LJ, Miller DG, Jacobs M, et al. Unique integration profiles in a canine model of long-term repopulating cells transduced with gammaretrovirus, lentivirus, and foamy virus. Hum Gene Ther. 2007;18:423–434. doi: 10.1089/hum.2007.011. [DOI] [PubMed] [Google Scholar]
- 14.Zufferey R, Donello JE, Trono D, Hope TJ. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kingsman SM, Mitrophanous K, Olsen JC. Potential oncogene activity of the woodchuck hepatitis post-transcriptional regulatory element (WPRE) Gene Ther. 2005;12:3–4. doi: 10.1038/sj.gt.3302417. [DOI] [PubMed] [Google Scholar]
- 16.Levesque JP, Winkler IG, Hendy J, Williams B, Helwani F, Barbier V, et al. Hematopoietic progenitor cell mobilization results in hypoxia with increased hypoxia-inducible transcription factor-1 alpha and vascular endothelial growth factor A in bone marrow. Stem Cells. 2007;25:1954–1965. doi: 10.1634/stemcells.2006-0688. [DOI] [PubMed] [Google Scholar]
- 17.Shima H, Takubo K, Iwasaki H, Yoshihara H, Gomei Y, Hosokawa K, et al. Reconstitution activity of hypoxic cultured human cord blood CD34-positive cells in NOG mice. Biochem Biophys Res Commun. 2009;378:467–472. doi: 10.1016/j.bbrc.2008.11.056. [DOI] [PubMed] [Google Scholar]
- 18.Alter BP, Knobloch ME, Weinberg RS. Erythropoiesis in Fanconi's anemia. Blood. 1991;78:602–608. [PubMed] [Google Scholar]
- 19.Waisfisz Q, Morgan NV, Savino M, de Winter JP, van Berkel CG, Hoatlin ME, et al. Spontaneous functional correction of homozygous fanconi anaemia alleles reveals novel mechanistic basis for reverse mosaicism. Nat Genet. 1999;22:379–383. doi: 10.1038/11956. [DOI] [PubMed] [Google Scholar]
- 20.Gregory JJ, Jr, Wagner JE, Verlander PC, Levran O, Batish SD, Eide CR, et al. Somatic mosaicism in Fanconi anemia: evidence of genotypic reversion in lymphohematopoietic stem cells. Proc Natl Acad Sci USA. 2001;98:2532–2537. doi: 10.1073/pnas.051609898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gross M, Hanenberg H, Lobitz S, Friedl R, Herterich S, Dietrich R, et al. Reverse mosaicism in Fanconi anemia: natural gene therapy via molecular self-correction. Cytogenet Genome Res. 2002;98:126–135. doi: 10.1159/000069805. [DOI] [PubMed] [Google Scholar]
- 22.Mankad A, Taniguchi T, Cox B, Akkari Y, Rathbun RK, Lucas L, et al. Natural gene therapy in monozygotic twins with Fanconi anemia. Blood. 2006;107:3084–3090. doi: 10.1182/blood-2005-07-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Shang X, Guo F, Murphy K, Kirby M, Kelley P, et al. Defective homing is associated with altered Cdc42 activity in cells from patients with Fanconi anemia group A. Blood. 2008;112:1683–1686. doi: 10.1182/blood-2008-03-147090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Follenzi A, Ailles LE, Bakovic S, Geuna M, Naldini L. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat Genet. 2000;25:217–222. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
- 26.Yamashita T, Kupfer GM, Naf D, Suliman A, Joenje H, Asano S, et al. The fanconi anemia pathway requires FAA phosphorylation and FAA/FAC nuclear accumulation. Proc Natl Acad Sci USA. 1998;95:13085–13090. doi: 10.1073/pnas.95.22.13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beard BC, Sud R, Keyser KA, Ironside C, Neff T, Gerull S, et al. Long-term polyclonal and multilineage engraftment of methylguanine methyltransferase P140K gene-modified dog hematopoietic cells in primary and secondary recipients. Blood. 2009;113:5094–5103. doi: 10.1182/blood-2008-09-176412. [DOI] [PMC free article] [PubMed] [Google Scholar]


