Abstract
Spatial priors, such as probabilistic atlases, play an important role in MRI segmentation. However, the availability of comprehensive, reliable and suitable manual segmentations for atlas construction is limited. We therefore propose a joint segmentation of corresponding, aligned structures in the entire population that does not require a probability atlas. Instead, a latent atlas, initialized by a single manual segmentation, is inferred from the evolving segmentations of the ensemble. The proposed method is based on probabilistic principles but is solved using partial differential equations (PDEs) and energy minimization criteria. We evaluate the method by segmenting 50 brain MR volumes. Segmentation accuracy for cortical and subcortical structures approaches the quality of state-of-the-art atlas-based segmentation results, suggesting that the latent atlas method is a reasonable alternative when existing atlases are not compatible with the data to be processed.
1 Introduction
Probabilistic atlases are crucial for most MR segmentation methods due to the absence of well defined boundaries. Derived from comprehensive sets of manually labeled examples, atlases provide statistical priors for tissue classification and structure segmentation [1,9,13,14,18]. Although atlas-based segmentation methods often achieve accurate results, the need for spatial priors can be problematic. First, the availability of suitable atlases is limited since manual segmentation of a significant number of volumes requires expensive effort of an experienced physician. Second, the suitability of existing atlases for images from different populations is questionable. Examples include using normal adult brain atlas for brain parcellation of young children or patients with severe brain pathologies.
Recently, a few methods have been proposed to reduce or avoid the dependency on possibly incompatible atlases. In the atlas-based segmentation method in [3], topological constraints are used to avoid possible bias introduced by the atlas. In [19], manually labeled structures are used to support the automatic segmentation of neighboring structures within the same image. Tu et al. [17] propose a discriminative approach for the segmentation of adjacent brain structures using a set of discriminative features learned from training examples. Lord et al. [11] suggest a group-wise smoothing, segmentation and registration method for cross sectional MR scans. Bhatia et al. [5] update an initial atlas constructed from a different population using the evolving segmentations of multiple images.
Here we propose and demonstrate a method that does not use a set of training images or probabilistic atlases as priors. Instead we extract an ensemble of corresponding structures simultaneously. The evolving segmentation of the entire image set supports each of the individual segmentations. In practice, a subset of the model parameters, called the spatial parameters, is inferred as part of the joint segmentation processes. These latent spatial parameters, which can be viewed as a ‘dynamic atlas’, are estimated exclusively from the data at hand and a single manual segmentation. The latent atlas is used as a Markov Random Field (MRF) prior on the tissue labels. The main novelty of the method with respect to other group-wise segmentation methods such as [11,5] is the consistent statistically-driven variational framework for MR ensemble segmentation.
Our contribution is two-fold. We introduce a level set framework, that is based on probabilistic principles, in which segmentation uncertainty is expressed by the logistic function of the associated level set values, similar to [15]. We then use it for group-wise segmentation. We evaluate our method by segmenting the amygdala, temporal gyrus and hippocampus in each hemisphere in 50 MR brain scans. The dice scores achieved by our method approach the atlas-based segmentation results of [14].
2 Problem Definition and Probabilistic Model
Our objective is to segment a particular structure or region of interest in N aligned MR images. Specifically, we consider the 2-partition problem where each voxel in image In (n = 1 … N) is assigned to either the foreground (structure of interest) or the background.
Let each image In: Ω → ℝ+, be a gray level image with V voxels, defined on Ω ⊂ ℝ3 and Γn: Ω → {0, 1} be the unknown segmentation of the image In. We assume that each Γn is generated iid from a probability distribution p(Γ| θΓ) where θΓ is a set of unknown parameters. We also assume that Γn generates the observed image In, independently of all other image-segmentation pairs, with probability p(In|Γn, θI,n) where θI,n are the parameters corresponding to image In. We assign a specific set of intensity parameters to each image since the acquisition conditions might vary across subjects.
Let {I1 … IN } be the given set of aligned images that form the observed variable in our problem and let Γ = {Γ1 … ΓN} be the corresponding segmentations. The joint distribution p(I1 … IN, Γ1 … ΓN|Θ) is governed by the composite set of parameters Θ = {θΓ, θI,1 … θI,N }. Our goal is to estimate the segmentations Γ.
We jointly optimize for the segmentations Γ and the parameters Θ, assuming that I1 … IN are independent:
| (1) |
| (2) |
| (3) |
We propose to alternate between estimating the maximum a posteriori (MAP) segmentations and updating the model parameters. For a given value of the model parameters Θ̂, Equation (3) implies that the segmentations can be estimated by solving N separate MAP problems:
| (4) |
We then fix Γ̂ and estimate the model parameters Θ = {θΓ, θI,1, … θI,N} by solving two ML problems:
| (5) |
| (6) |
In the following sections we present a level-set framework that is motivated by this probabilistic model. We reformulate the estimation problem stated in Eq. (4) such that the soft segmentations p(Γn) rather then the Γn are estimated.
3 Probabilistic View of the Level Set Framework
Now we draw the connection between the probabilistic model presented above and the level set framework for segmentation. Let φn: Ω → ℝ denote a level set function associated with image In. The zero level Cn = {x ∈ Ω| φn(x) = 0} defines the interface between the partitions of In. Vese and Chan [6] represent the image partitions by a regularized variant of the Heaviside function of φn, e.g., . Alternatively, we can use the hyperbolic tangent to achieve the same goal:
| (7) |
For ε = 1, the function H̃ε(·) is the logistic function. Similar to [15], we define the level set function φn using the log-odds formulation instead of the conventional signed distance function:
| (8) |
The scalar ε determines the scaling of the level set function φn with respect to the ratio of the probabilities. Substituting this definition into Eq. (7) we obtain
| (9) |
which implies that the function H̃ε(φn(x)) can be viewed as the probability that the voxel in location x belongs to the foreground region. The functions H̃ε(φn(x)) and H(φn(x)) therefore represent soft and hard segmentations, respectively. To simplify the notation we omit the subscript ε in the rest of the paper.
In the following subsections we relate the terms in Eq. (3) to the energy terms in the classical level set functional.
3.1 Image Likelihood Term
Let us first consider the image likelihood term in Eq. (3):
| (10) |
where pin and pout are the probability distributions of the foreground and back-ground image intensities, respectively.
Let EI ≅ −log p(In| Γn, θ̂I,n) define the energy term associated with the image likelihood term. Using the level-set formulation and replacing the binary labels Γn in Eq. (10) with a soft segmentation represented by H̃(φn), we get:
| (11) |
If we use, for example, Gaussian densities for pin and pout we get the familiar minimal variance term [6,12]. Here, we use a Gaussian mixture to model the background, as described later in the paper.
3.2 Spatial Prior Term
We define the prior probability p(Γn|θΓ) to be a Markov Random Field (MRF):
| (12) |
where Z(θΓ) is the partition function and  (v) are the closest neighbors of voxel v. The function f (·) accounts for the interactions between neighboring voxels. If we omit the pairwise term in Eq. (12), the prior on segmentations p(Γn|θΓ) reduces to a Bernoulli distribution, where the parameters θΓ represent the probability map for the structure of interest. The introduction of the pairwise clique potentials complicates the model but encourages smoother labeling configurations.
(v) are the closest neighbors of voxel v. The function f (·) accounts for the interactions between neighboring voxels. If we omit the pairwise term in Eq. (12), the prior on segmentations p(Γn|θΓ) reduces to a Bernoulli distribution, where the parameters θΓ represent the probability map for the structure of interest. The introduction of the pairwise clique potentials complicates the model but encourages smoother labeling configurations.
Using the continuous level set formulation with soft segmentation, we define the spatial energy term. as follows:
| (13) |
As in [4] we ignore the partition function, approximating the MRF model above. The logarithm of the pairwise clique potential term f (·) can be configured to act as a finite difference operator approximating the gradient of Γn at the voxel v [10]. It can be therefore viewed as an approximation of the continuous term
| (14) |
which is the commonly used smoothness constraint as reformulated in [6].
3.3 The Unified Energy Functional
We now construct the cost functional for φ1 … φN and the parameters Θ by combing Eqs. (11),(13) and (14):
| (15) |
where α = 1 − β − γ. As in [16] we tune the weights such that the contributions of the energy terms ELEN, EI and ES to the overall cost are balanced.
4 Alternating Minimization Algorithm
We optimize the unified functional (15) in a set of alternating steps. For fixed model parameters Θ, the evolution of each of the level set functions φn follows the gradient descent equation:
| (16) |
where is the derivative of the Heaviside function, i.e.,
For fixed segmentations φn the model parameters are recovered by differentiating the cost functional (15) with respect to each parameter.
4.1 Intensity Parameters
We assume that the intensities of the structure of interest are drawn from a normal distribution, i.e., . The distribution mean and variance of the foreground region of In are updated at every iteration. The intensities of the background tissues are modeled as a K-Gaussian mixture:
where is the mixing proportion component k in the mixture. The Gaussian mixture model parameters are estimated using expectation maximization (EM) [7].
4.2 Spatial Parameters
We estimate the spatial function θΓ(x), constructing a dynamically evolving latent atlas, by optimizing the sum of the energy terms the depend on θΓ:
yielding .
5 Experimental Results
We test the proposed approach on 50 MR brain scans. Some of the subjects in this set are diagnosed with the first episode schizophrenia or affective disorder. The MR images (T1, 256 × 256 × 128 volume, 0.9375 × 0.9375 × 1.5 mm3 voxel size) were acquired by a 1.5-T General Electric Scanner. The data was originally acquired for brain morphometry study [8]. In addition to the MR volumes, manual segmentations of three structures (superior temporal gyrus, amygdala, and hippocampus) in each hemisphere were provided for each of the 50 individuals and used to evaluate the quality of the automatic segmentation results. MR images are preprocessed by skull stripping. The volumes were aligned using B-spline registration according to [2].
Assuming that the manual segmentation of a single instance is given, we initialize the latent atlas θΓ by the Heaviside function of a single manual segmentation smoothed with a Gaussian kernel of width σ (σ = .35 in our experiments). We ran the experiments twice, initializing the level-set functions of the signed distance function of the given manual segmentation or a sphere, with center and radius corresponding to the manually segmented structure. The results obtained using the first mode of initialization were slightly better. We excluded the image associated with the given manual segmentations from the ensemble. The algorithm was implemented in Matlab and ran on the average 7.5 minutes for 50 cropped volumes, excluding the time of the initial estimate of the background intensity of the Gaussian mixture. About 7 iterations were needed until convergence which was obtained when the update of the level-set functions did not induce changes in the corresponding boundaries.
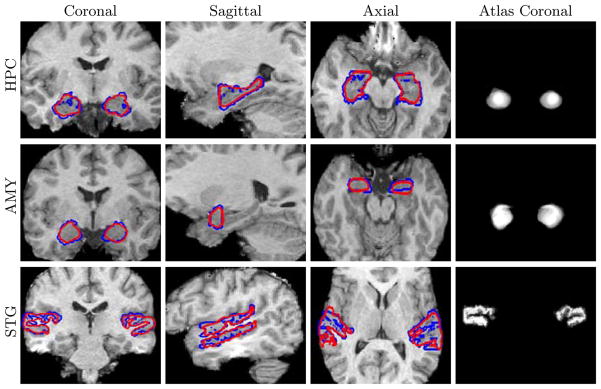
We used the Dice score to evaluate segmentations. The results are shown in Figure 1. In contrast to [14] we do not use spatial priors, neither do we use hierarchical multi-stage segmentation model. Nevertheless, the Dice scores obtained by our method approache the state-of-the-art atlas-based segmentation results reported there. To exemplify the significance of a latent atlas, generated concurrently with the segmentation, we also show a comparison to the segmentation results obtained by using an atlas constructed from a single manual segmentation smoothed by a Gaussian kernel, without the update procedure. Figure 2 shows segmentation examples of the three pairs of structures in representative individual brains. Coronal views of the resulting 3D atlases for each pair of structures are shown in the fourth column of Figure 2.
Fig. 1.
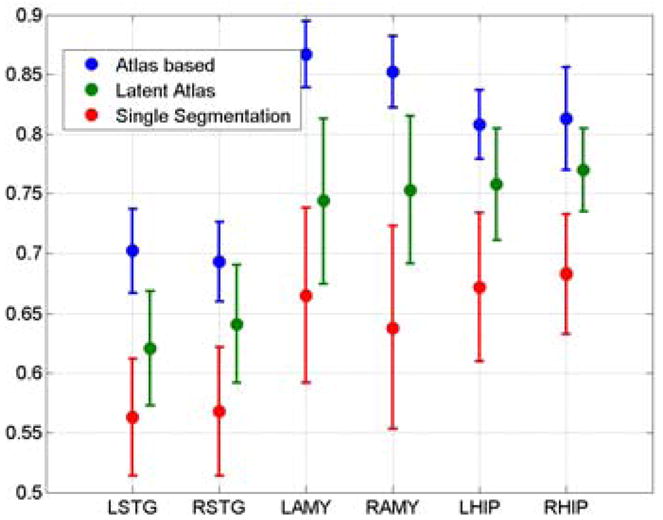
The mean and standard deviation of the Dice scores calculated for six structures in the ensemble. The latent atlas segmentation (green) is compared to the atlas-based segmentation (blue) reported in [14] and to the segmentation obtained by using a single manual segmentation as an atlas (red).
Fig. 2.
Three cross-sections of 3D segmentations of Hippocampus, Amygdala and Superior Temporal Gyrus in the left and right hemispheres. Automatic segmentation is shown in red. Manual segmentation is shown in blue. Fourth column: Coronal views of the resulting atlases for each pair of structures.
6 Discussion and Conclusions
We presented a level set framework for segmentation of MR image ensembles, that is motivated by a generative probabilistic model. Unlike most previous methods, we do not use spatial priors in the form of a probabilistic atlas. Instead, spatial latent parameters, which form a ‘dynamic atlas’, are inferred from the data set through an alternating minimization procedure.
The quality of the segmentation results obtained for ensembles of brain structures shows that the proposed method presents a reasonable alternative to standard segmentation techniques when a compatible atlas is not available.
An on-going research is now conducted to demonstrate the ability of the proposed algorithm to handle pathological cases (e.g., in a longitudinal or a multimodal study) where the atlas-based approach still fails.
Acknowledgments
This work was supported in part by NIH NIBIB NAMIC U54-EB005149, NIH NCRR NAC P41-RR13218, NIH NINDS R01-NS051826, NIH NCRR mBIRN U24-RR021382 grants and NSF CAREER Award 0642971.
References
- 1.Ashburner J, Friston K. Unified segmentation. NeuroImage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 2.Balci SK, Golland P, Shenton M, Wells WM. Free-form B-spline deformation model for groupwise registration. Proc. of MICCAI 2007 Statistical Registration Workshop: Pair-wise and Group-wise Alignment and Atlas Formation; 2007. pp. 23–30. [Google Scholar]
- 3.Bazin P-L, Pham DL. Statistical and topological atlas based brain image segmentation. In: Ayache N, Ourselin S, Maeder A, editors. MICCAI 2007, Part I. LNCS. Vol. 4791. Springer; Heidelberg: 2007. pp. 94–101. [DOI] [PubMed] [Google Scholar]
- 4.Besag J. Statistical analysis of non-lattice data. The Statistician. 1975;24(3):179–195. [Google Scholar]
- 5.Bhatia KK, Aljabar P, Boardman JP, Srinivasan L, Murgasova M, Counsell SJ, Rutherford MA, Hajnal JV, Edwards AD, Rueckert D. Groupwise combined segmentation and registration for atlas construction. In: Ayache N, Ourselin S, Maeder A, editors. MICCAI 2007, Part I. LNCS. Vol. 4791. Springer; Heidelberg: 2007. pp. 532–540. [DOI] [PubMed] [Google Scholar]
- 6.Chan T, Vese L. Active contours without edges. TIP. 2001;10(2):266–277. doi: 10.1109/83.902291. [DOI] [PubMed] [Google Scholar]
- 7.Dempster A, Laird N, Rubin D. Maximal likelihood form incomplete data via the EM algorithm. Proc of the Royal Statistical Society. 1977;39:1–38. [Google Scholar]
- 8.Hirayasu Y, Shenton ME, Salisbury DF, Dickey CC, Fischer IA, Mazzoni P, Kisler T, Arakaki H, Kwon JS, Anderson JE, Yurgelun-Todd D, Tohen M, McCarley RW. Lower left temporal lobe MRI volumes in patients with first-episode schizophrenia compared with psychotic patients with first-episode affective disorder and normal subjects. Amer J Psychiatry. 1998;155(10):1384–1391. doi: 10.1176/ajp.155.10.1384. [DOI] [PubMed] [Google Scholar]
- 9.Fischl B, Salat D, Busa E, Albert M, Dietrich M, Haselgrove C, Van Der Kouwe A, Killany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale A. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 10.Li S. Markov Random Field Modeling in Computer Vision. Springer; Heidelberg: 1995. [Google Scholar]
- 11.Lord N, Ho J, Vemuri B. ICCV. 2007. Ussr: A unified framework for simultaneous smoothing, segmentation, and registration of multiple images; pp. 1–6. [Google Scholar]
- 12.Paragios N, Deriche R. Geodesic active regions: A new paradigm to deal with frame partition problems in computer vision. JVCIR. 2002;13:249–268. [Google Scholar]
- 13.Pohl KM, Fisher J, Grimson WEL, Kikinis R, Wells WM. A bayesian model for joint segmentation and registration. NeuroImage. 2006;31(1):228–239. doi: 10.1016/j.neuroimage.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 14.Pohl KM, Bouix S, Nakamura M, Rohlfing T, McCarley RW, Kikinis R, Grimson WEL, Shenton ME, Wells WM. A hierarchical algorithm for MR brain image parcellation. TMI. 2007;26(9):1201–1212. doi: 10.1109/TMI.2007.901433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pohl KM, Fisher J, Bouix S, Shenton M, McCarley R, Grimson WEL, Kikinis R, Wells WM. Using the logarithm of odds to define a vector space on probabilistic atlases. Medical Image Analysis. 2007;11(6):465–477. doi: 10.1016/j.media.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riklin-Raviv T, Sochen N, Kiryati N. Shape-based mutual segmentation. International Journal of Computer Vision. 2008;79:231–245. [Google Scholar]
- 17.Tu Z, Narr KL, Dollár P, Dinov I, Thompson PM, Toga AW. Brain anatomical structure segmentation by hybrid discriminative/generative models. IEEE TMI. 2008;27(4):495–508. doi: 10.1109/TMI.2007.908121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Leemput K, Maes F, Vandermeulen D, Suetens P. Automated model-based tissue classification of MR images of the brain. IEEE TMI. 1999;18(10):897–908. doi: 10.1109/42.811270. [DOI] [PubMed] [Google Scholar]
- 19.Yang J, Duncan J. CVPR. 2004. Joint prior models of neighboring object for 3D image segmentation; pp. 314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]