Abstract
Objectives
To directly assess whether or not genome-wide expression profiles derived from leukocyte subsets are comparable to that of whole blood, as measured by enrichment for genes corresponding to metabolic and signaling pathways.
Design
Prospective observational study involving microarray-based bioinformatics based on RNA individually derived from whole blood, neutrophils, monocytes, and lymphocytes, respectively.
Setting
Three pediatric intensive care units (PICU) in the United States.
Patients
Children ≤ 10 years of age: 5 normal controls and 13 meeting criteria for septic shock on day 1 of presentation to the PICU.
Interventions
None other than standard care.
Measurements and Main Results
Baseline analyses using whole blood-derived RNA demonstrated increased expression of genes corresponding to signaling pathways involving innate immunity, redox balance, and protein ubiquitination, and decreased expression of genes corresponding to the adaptive immune system. Subsequent analyses using leukocyte-specific RNA were congruent with the gene expression profiles demonstrated using whole blood-derived RNA, as measured by enrichment for genes corresponding to metabolic and signaling pathways. Gene network analysis, derived from a composite gene list involving the individual gene expression profiles of neutrophils, monocytes, and lymphocytes, respectively, revealed a gene network corresponding to antigen presentation, cell-mediated immunity, and humoral-mediated immunity. Finally, a sub-analysis focused on network gene nodes localized to the nuclear compartment revealed functional annotations related to transcriptional repression and epigenetic regulation.
Conclusions
These data demonstrate that genome-level repression of adaptive immunity gene programs early in the course of pediatric septic shock remained evident when analyses are conducted using leukocyte subset-specific RNA.
Keywords: microarray, T cell, antigen presentation, children, inflammation
INTRODUCTION
We have recently published a series of manuscripts involving genome-wide expression profiling in pediatric septic shock [1–4]. These translational studies have served to generate novel hypotheses at the genomic level and to identify disease and illness severity biomarkers [5, 6]. All of these genome-wide expression studies have been based on whole blood derived-RNA. Because whole blood-derived RNA represents a mixed population of blood cells, a potentially valid and important criticism of these previous studies is that they are confounded by the differential white blood cell counts of the individual study subjects.
We have indirectly addressed this important criticism through a series of validation studies indicating that our previous observations are not merely epiphenomena of differential white blood cell counts [1, 3, 4]. Herein, we seek to directly address this issue by conducting genome-wide expression studies using RNA specifically derived from the 3 major leukocyte subpopulations: neutrophils, monoyctes, and lymphocytes. Thus, the major objective of this study is to directly assess whether or not genome-wide expression patterns derived from leukocyte subsets are comparable to that of whole blood. The primary approach for assessing comparability will be the coordinated regulation of genes that correspond to established metabolic and signaling pathways. Correspondence to established metabolic and signaling pathways will be objectively measured by uploading gene lists to the Ingenuity Pathways Analysis application as previously described [1, 3, 7].
METHODS
Patients
The study protocol was approved by the individual Institutional Review Boards of each participating institution. Children ≤ 10 years of age admitted to the PICU and meeting published, pediatric-specific criteria for septic shock were eligible [8]. Control patients were recruited from the ambulatory departments of participating institutions using previously published inclusion / exclusion criteria [2].
Sample and data collection
After obtaining informed consent from parents or legal guardians, blood samples were obtained within the first 24 hours of meeting criteria for septic shock. Severity of illness was calculated using the PRISM III score [9]. Organ failure was defined using pediatric-specific criteria [8]. Annotated clinical and laboratory data were collected daily while in the PICU using a web-based database developed locally.
RNA extraction and microarray hybridization
The data and protocols described in this manuscript are MIAME compliant are in the process of being deposited in the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/).
Total RNA was isolated from whole blood samples using the PaxGene™ Blood RNA System (PreAnalytiX, Qiagen/Becton Dickson, Valencia, CA) according the manufacturer’s specifications. When using the PAXGene™ system an aliquot of whole blood is immediately placed into a collection tube containing a proprietary solution that lyses all blood cell components and stabilizes RNA. The resulting suspension is then subjected to a series of steps to isolate RNA from the whole blood samples. A separate aliquot of whole blood was used to isolate leukocyte subsets. Individual leukocytes subsets were first isolated using the Miltenyi autoMACS Cell Separation platform and MACS Microbeads targeted at specific isolation of neutrophils, monocytes, and lymphocytes, and according the manufacturer’s specifications (Miltenyi Biotec Inc., Auburn, CA). We typically achieve leukocyte subset purity of >96% using this protocol (data not shown). After leukocyte subset isolation, leukocyte subset total RNA was isolated using the RNeasy RNA Isolation Kit (Qiagen, Valencia, CA). Microarray hybridization was performed by the Affymetrix Gene Chip Core facility at Cincinnati Children’s Hospital Research Foundation as previously described using the GeneChip Human Gene 1.0 Array (Affymetrix, Santa Clara, CA) [1–4].
Data analysis
Analyses were performed using one patient sample per RNA sample. Image files were captured using an Affymetrix GeneChip Scanner 3000. CEL files were subsequently preprocessed using Robust Multiple-Array Average (RMA) normalization using GeneSpring GX 7.3 software (Agilent Technologies, Palo Alto, CA). All signal intensity-based data were used after RMA normalization, which specifically suppresses all but significant variation among lower intensity probe sets [10]. All chips representing patient samples and RNA source were then normalized to the respective median values of controls.
Differences in mRNA abundance between the septic shock patients and controls, for each type of RNA, were measured by sequential expression and statistical filters using GeneSpring GX 7.3. The expression filter was applied first and used all gene probes on the array (>30,000). We selected only the genes having a least 1.5-fold expression difference between the medians of controls and patients with septic shock, for each RNA type. The genes that passed this initial expression filter were then subjected to a statistical filter consisting of ANOVA (Wilcoxon) using the septic shock patients and controls as the comparison groups, and corrections for multiple comparisons based on a Benjamini-Hochberg false discovery rate of 5% [11].
Gene lists of differentially expressed genes were primarily analyzed using the Ingenuity Pathways Analysis application (IPA, Ingenuity Systems, Redwood City, CA) that provides a tool for discovery of signaling pathways and gene networks within the uploaded gene lists as previously described [1, 7]. Adjunct analyses of gene lists were performed using ToppGene, a distinct, public, relational database of functional gene annotations [12]. These applications are all based on the established biomedical literature and use specific approaches to estimate significance (p values) based on non-redundant representations of the microarray chip and to convert the uploaded gene lists to gene lists containing a single value per gene. The p values provide an estimate of the probability that a given enrichment is present by chance alone and are derived using corrections for multiple comparisons.
RESULTS
Basic demographic data and whole blood-derived gene expression patterns
Table 1 provides general clinical data for each patient in the septic shock cohort (n = 13). There were no significant differences between the control cohort (n = 5) and the septic shock cohort with respect to age and gender (data not shown). The other demographic variables for the septic shock cohort provided in Table 1 are consistent with our previous reports [1–4].
Table 1.
General clinical information for the septic shock cohort.
| Pt. # |
Gender | Race | Age (yrs) |
Survivor | PRISM | Organism (site) | Other Diagnoses |
|---|---|---|---|---|---|---|---|
| 1 | Male | White | 0.5 | Yes | 6 | None | Peritonitis |
| 2 | Male | African- American |
0.9 | Yes | 7 | Influenza A (lung) | Necrotizing pneumonia |
| 3 | Female | White | 2.4 | Yes | 5 |
P. aeruginosa (lung) |
Small bowel perforation |
| 4 | Female | White | 9.1 | Yes | 16 |
P. aeruginosa (lung) |
Developmental delay |
| 5 | Female | White | 9.6 | No | 25 | None | Epilepsy |
| 6 | Male | White | 5.3 | Yes | 15 |
S. pneumoniae (lung) |
Developmental delay |
| 7 | Male | African- American |
3.7 | Yes | 14 | E. coli (blood) | Short gut syndrome |
| 8 | Male | White | 3 | Yes | 11 |
S. pneumoniae (lung) |
Necrotizing pneumonia |
| 9 | Male | White | 0.4 | Yes | 9 | None | Pneumonia |
| 10 | Male | White | 1.3 | Yes | 35 |
S. pneumoniae (lung) |
Prader-Willi, pneumonia |
| 11 | Male | African- American |
4.4 | Yes | 15 | None | Pneumonia |
| 12 | Male | White | 0.6 | No | 23 |
K. pneumoniae (blood) |
Short gut syndrome |
| 13 | Female | White | 8.1 | Yes | 12 |
S. pyogenes (blood) |
None |
To begin addressing the primary objective of the study, we first derived a list of genes differentially regulated between controls and septic shock patients using whole blood-derived RNA. Based on whole blood-derived RNA there were 2,355 differentially regulated genes in the septic shock patients, relative to controls (1,283 upregulated and 1,072 downregulated). Neutrophils typically outnumber monocytes and lymphocytes in whole blood and therefore have the potential to be over-represented in genome-wide expression analyses. To investigate this possibility, we searched the 2,355 gene list for the presence of “signature probe sets” for neutrophils, monocytes, and lymphocytes, respectively [13]. In the 2,355 gene list there were 7 neutrophil signature probe sets, 6 monocyte signature probe sets, and 20 lymphocyte signature probe sets, thus demonstrating that all major leukocyte subsets are well represented in the 2,355 gene list.
The 1,283 upregulated gene list and the 1,072 downregulated gene list were individually uploaded to the IPA application and the analytical output was focused on enrichment for signaling and metabolic pathways. The IPA application analyzes a given gene list, and based on the established biomedical literature, objectively determines if the gene list is significantly enriched for known metabolic and signaling pathways. Significance is based on a p value that provides an estimate of the probability that a given enrichment is present by chance alone. The strength of the p value is directly proportional to the number of genes in the list that correspond to a given pathway, and indirectly proportional to the total number of genes in the entire gene list.
Table 2A provides the top 10 (based on p values) signaling and metabolic pathways represented by the 1,283 upregulated genes. These upregulated pathways globally represent processes related to innate immunity, redox balance, and protein ubiquitination. Table 2B provides the top 10 (based on p values) signaling and metabolic pathways represented by the 1,072 downregulated genes. These downregulated pathways primarily correspond to the adaptive immune system. Thus, in this cohort of septic shock patients, the genome-wide expression pattern based on whole blood-derived RNA is most notable for repression of genes corresponding to signaling pathways of the adaptive immune system. This observation is consistent with our previous reports and provides a reference point for our subsequent analyses focused on leukocyte subset-specific gene expression profiles in the same patient cohort.
Table 2.
| A: Top 10 signaling and metabolic pathways represented by the 1,283 upregulated genes based on whole blood-derived RNA. | ||
|---|---|---|
| PATHWAY | P VALUE | # OF GENES |
| Oxidative phosphorylation | 8.3E-17 | 36 |
| Mitochondrial dysfunction | 2.7E-10 | 27 |
| IL-10 signaling | 2.5E-7 | 15 |
| Toll-like receptor signaling | 4.0E-6 | 12 |
| NRF2-mediated oxidative stress response | 7.2E-6 | 24 |
| Ubiquinone biosynthesis | 4.5E-5 | 13 |
| TREM1 signaling | 7.9E-5 | 11 |
| NF-κB signaling | 1.9E-4 | 18 |
| Protein ubiquitination pathway | 3.3E-4 | 20 |
| IL-6 signaling | 6.7E-4 | 13 |
| B: Top 10 signaling and metabolic pathways represented by the 1,072 downregulated genes based on whole blood-derived RNA. | ||
|---|---|---|
| PATHWAY | P VALUE | # OF GENES |
| iCOS signaling in T helper cells | 7.0E-19 | 27 |
| Calcium-induced T lymphocyte apoptosis | 5.4E-14 | 18 |
| CD28 signaling in T helper cells | 1.4E-13 | 23 |
| Role of NFAT in regulation of the immune response | 4.1E-13 | 28 |
| T cell receptor signaling | 1.8E-12 | 20 |
| CTLA4 signaling in cytotoxic T lymphocytes | 3.8E-8 | 15 |
| CCR5 signaling in macrophages | 1.2E-6 | 12 |
| Cytotoxic T lymphocyte-mediated apoptosis | 2.6E-6 | 8 |
| Allograft rejection signaling | 3.7E-6 | 9 |
| T helper cell differentiation | 5.8E-6 | 9 |
Global gene list comparisons based on leukocyte subset-specific RNA
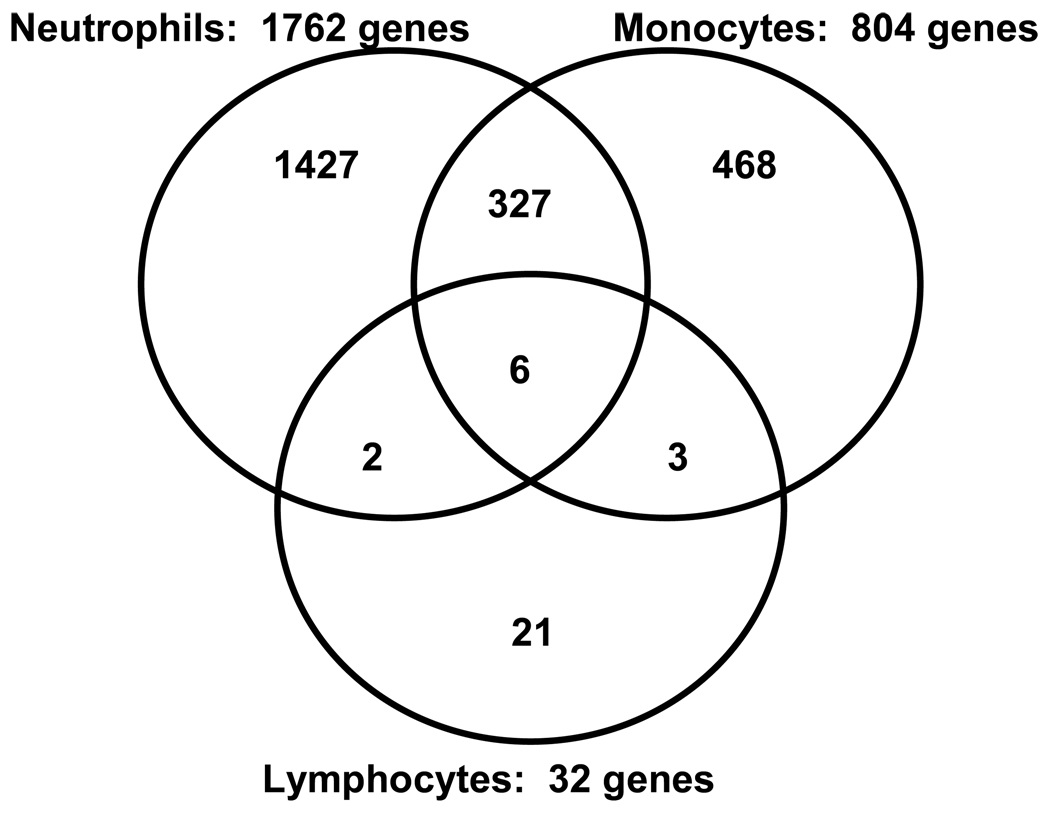
In the next set of analyses we conducted genome-wide expression profiling using RNA from specific leukocyte subsets (neutrophils, monocytes, and lymphocytes, respectively). Figure 1 depicts the number of upregulated genes in patients with septic shock, relative to controls, for each leukocyte subset. The largest number of upregulated genes was observed in neutrophils, followed by monocytes and lymphocytes, respectively. Venn analysis demonstrated that for each leukocyte subset, the majority of upregulated genes (>58%) were unique to a given leukocyte subset. The largest overlap of commonly upregulated genes was observed between neutrophils and monocytes.
Figure 1.
Venn analysis comparing the genes differentially upregulated between patients with septic shock and controls for each leukocyte subset.
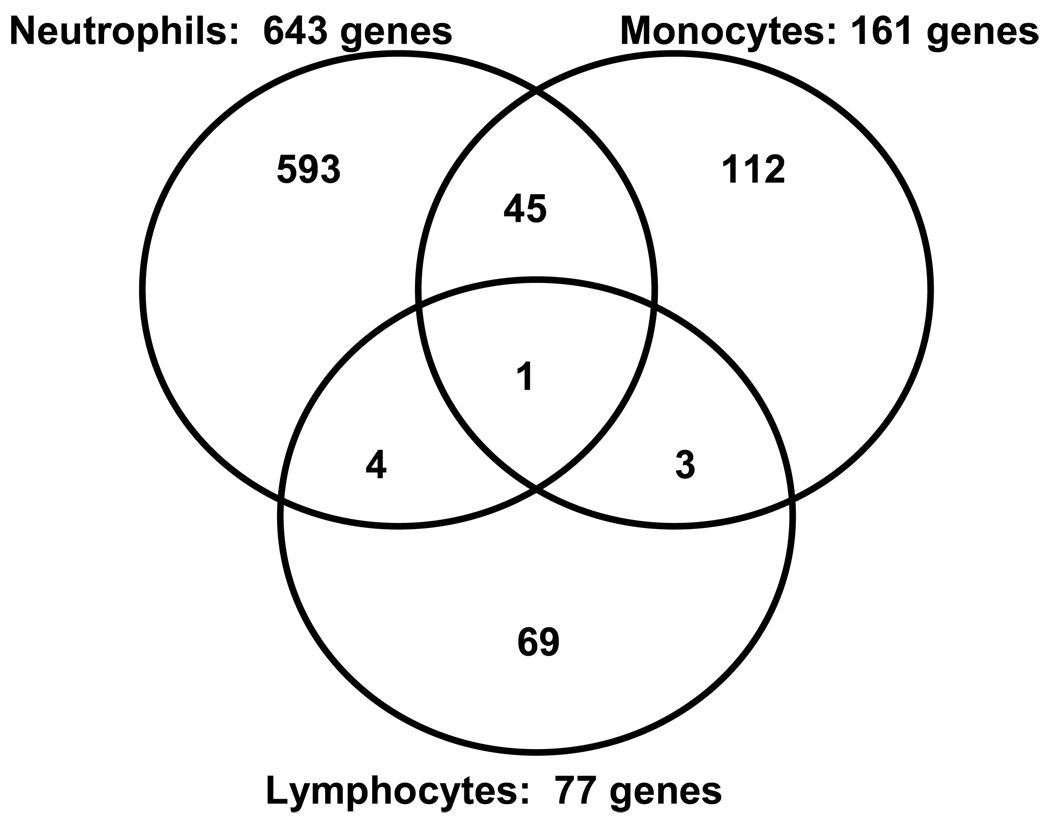
Figure 2 depicts the number of downregulated genes in patients with septic shock, relative to controls, for each leukocyte subset. The largest number of downregulated genes was observed in neutrophils, followed by monocytes and lymphocytes, respectively. Venn analysis demonstrated that for each leukocyte subset, the majority of downregulated genes (>69%) were unique to a given subset. The largest overlap of commonly downregulated genes was observed between neutrophils and monocytes.
Figure 2.
Venn analysis comparing the genes differentially downregulated between patients with septic shock and controls for each leukocyte subset.
These data indicate that the three major leukocyte subsets have relatively unique patterns of differential gene expression in septic shock patients. The data also indicate that neutrophils have the greatest degree of differential gene expression, while lymphocytes have the least degree of differential gene expression.
Pathway analysis for upregulated genes in specific leukocyte subsets
In the next set of analyses we sought to derive more functional information from the leukocyte subset-specific gene lists described above. Accordingly, individual lists of upregulated genes, for each leukocyte subset, were uploaded to the IPA application and the analytical output was focused on enrichment for genes corresponding to signaling and metabolic pathways. Tables 3A and 3B provide the top five (based on p values) signaling and metabolic pathways represented by the upregulated genes in neutrophils and monocytes, respectively (pathway specific genes are listed in the Supplementary Data). The list of genes upregulated in lymphocytes was not significantly enriched for any signaling or metabolic pathways. Table 3A demonstrates that the list of upregulated neutrophil genes was most enriched for genes corresponding to the protein ubiquitination pathway. In addition, the upregulated neutrophil genes were enriched for pathways related to mitochondrial dysfunction and redox balance-related signaling. Table 2B demonstrates that the list of upregulated monocyte genes was most enriched for caveolar-mediated endocytosis. In addition, the upregulated monocyte genes were enriched for the interleukin (IL)-10 and IL-12 signaling pathways.
Table 3.
| A: Top signaling and metabolic pathways represented by the 1,762 upregulated genes based on neutrophil-derived RNA | ||
|---|---|---|
| PATHWAY | P VALUE | # OF GENES |
| Protein ubiquitination pathway | 6.8E-6 | 32 |
| NRF2-mediated oxidative stress response | 5.5E-5 | 27 |
| Mitochondrial dysfunction | 7.1E-5 | 22 |
| Oxidative phosphorylation | 1.4E-4 | 22 |
| PI3K/Akt signaling | 4.8E-3 | 16 |
| B: Top signaling and metabolic pathways represented by the 804 upregulated genes based on monocyte-derived RNA. | ||
|---|---|---|
| PATHWAY | P VALUE | # OF GENES |
| Calveolar-mediated endocytosis | 9.6E-7 | 13 |
| NRF2-mediated oxidative stress response | 4.0E-5 | 18 |
| IL-10 signaling | 2.5E-4 | 9 |
| Leukocyte extravasation signaling | 3.3E-3 | 14 |
| IL-12 signaling in macrophages | 4.3E-3 | 10 |
Collectively, these data demonstrate that the neutrophil and monocyte genes upregulated in septic shock correspond to distinct signaling pathways relevant to the pathobiology of septic shock. In addition, the signaling and metabolic pathways provided in Tables 3A and 3B are in concordance with the data generated using whole-blood-derived RNA. Finally, the data also suggest that the upregulated lymphocyte genes are not particularly enriched for any specific signaling or metabolic pathway.
Pathway analysis for downregulated genes
Individual lists of downregulated genes, for each leukocyte subset, were uploaded to the IPA application and the analytical output was again focused on enrichment for genes corresponding to signaling and metabolic pathways. Tables 4A – 4C provide the top five (based on p values) signaling and metabolic pathways represented by the downregulated genes in neutrophils, monocytes, and lymphocytes, respectively (pathway specific genes are listed in the Supplementary Data). Table 4A demonstrates that the list of downregulated neutrophil genes was most enriched for genes corresponding to the antigen presentation pathway and interferon signaling. Table 4B demonstrates that the list of downregulated monocyte genes was most enriched for signaling pathways related to the antigen presentation pathway, dendritic cell maturation, and modulation of T cell activity. Table 4C demonstrates that the list of downregulated lymphocyte genes was most enriched for pathways relevant to natural killer cell signaling and T cell function. These data are also in concordance with the whole blood-derived RNA data, and demonstrate that the neutrophil, monocyte, and lymphocyte genes downregulated in septic shock correspond to distinct signaling pathways particularly relevant to the adaptive immune system.
Table 4.
| A: Top signaling and metabolic pathways represented in the 643 downregulated genes from neutrophil-derived RNA. | ||
|---|---|---|
| PATHWAY | P VALUE | # OF GENES |
| Antigen presentation pathway | 3.1E-9 | 10 |
| Interferon signaling | 5.4E-5 | 6 |
| Hepatocyte growth factor signaling | 2.5E-4 | 9 |
| Pattern recognition receptor signaling | 5.5E-4 | 8 |
| Eicosanoid signaling | 5.6E-4 | 7 |
| B: Top signaling and metabolic pathways represented in the 161 downregulated genes from monocyte-derived RNA. | ||
|---|---|---|
| PATHWAY | P VALUE | # OF GENES |
| Antigen presentation pathway | 4.2E-9 | 6 |
| Dendritic cell maturation | 2.9E-5 | 6 |
| IL-4 signaling | 1.5E-4 | 4 |
| CTLA4 signaling in cytotoxic T lymphocytes | 3.6E-4 | 4 |
| Allograft rejection signaling | 4.2E-4 | 3 |
| C: Top signaling and metabolic pathways represented in the 77 downregulated genes from lymphocyte-derived RNA. | ||
|---|---|---|
| PATHWAY | P VALUE | # OF GENES |
| NK cell signaling | 3.7E-7 | 6 |
| iCOS signaling in T helper cells | 8.0E-6 | 6 |
| CD28 signaling in T helper cells | 1.3E-5 | 6 |
| Calcium-induced T lymphocyte apoptosis | 1.7E-5 | 5 |
| CTLA4 signaling in cytotoxic T lymphocytes | 8.0E-5 | 5 |
Network analysis
In the previous analyses we examined each leukocyte subset expression pattern in isolation. Herein, we tested the hypothesis that the individual leukocyte subset expression patterns, when analyzed in combination, can contribute to coordinately regulated gene networks. To this end we extracted the genes common to all gene lists represented by the Venn diagrams in Figure 1 and Figure 2 (n = 3,072 genes) and uploaded this entire merged gene list to the IPA application. The output of this analysis was focused on enrichment for gene networks.
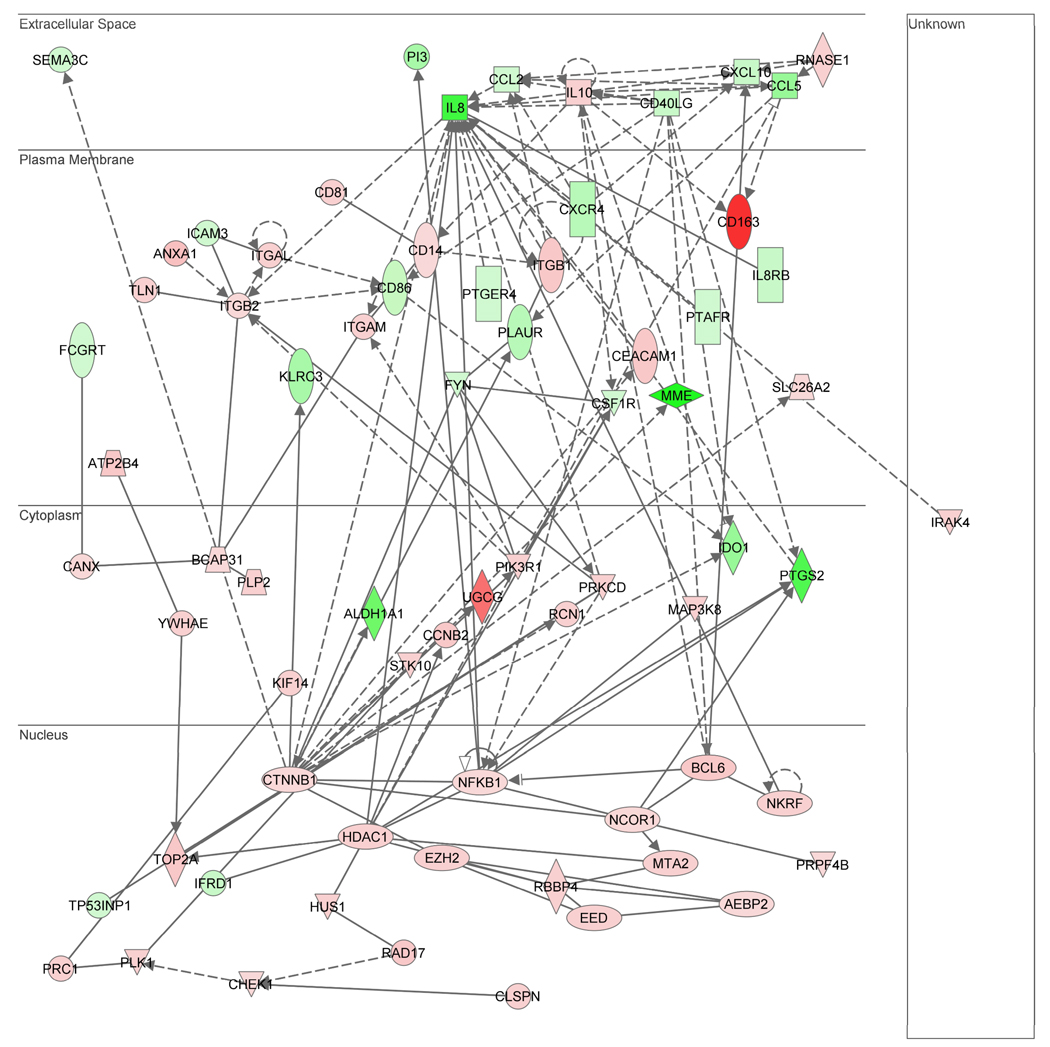
The highest scoring (i.e. most statistically significant) gene network represented in this merged gene list is shown in Figure 3, with red intensity indicating increased expression and green intensity indicating decreased expression in patients with septic shock, relative to controls. The gene network consists of 70 gene nodes and is depicted based on cellular compartment localization of the individual gene nodes. The network contains IL-8 as a highly connected gene node in the extracellular compartment and β-catenin (CTNNB1) as a highly connected gene node in the nuclear compartment. The top functional annotations associated with this network (based on the IPA algorithm) are antigen presentation, cell-mediated immune response, and humoral immune response. Among the 70 genes in the network, 36 (51%) were derived from neutrophils, 20 (29%) were derived from monocytes, and 4 (6%) were derived from lymphocytes. Of the remaining 10 genes, 9 genes were common to both neutrophils and monocytes, and 1 gene was common to both monocytes and lymphocytes. These data demonstrate that each of the leukocyte subset-specific gene expression profiles contribute to a gene network enriched for functional annotations particularly relevant to the adaptive immune system.
Figure 3.
Gene network derived from a merged list of genes common to all gene lists represented by the Venn diagrams in Figure 1 and Figure 2. Red intensity within in a gene node corresponds to increased expression in the patients with septic shock, relative to controls, and green intensity within a gene node corresponds to decreased expression in the patients with septic shock, relative to controls. This network has a score of 28, which is equivalent to a p value of 1.0E-28. The p value provides an estimate of the probability that the network genes are present in the uploaded gene list by chance alone. A network legend and a complete list of the network genes are provided in the Supplementary Data.
Network genes in the nuclear compartment
The gene network depicted in Figure 3 demonstrates 21 gene nodes in nuclear compartment, and the majority of these genes (19 of 21) are upregulated in the septic shock patients relative to controls. Based on these observations we uploaded the 21 gene nodes in the nuclear compartment to a functional annotation database independent of IPA (ToppGene) and the analytical output was focused on enrichment for molecular function annotations. Table 5 provides the top 5 (based on p values) molecular function annotations for the 21 gene nodes in the nuclear compartment. These molecular function annotations are consistent with repression of transcriptional activity via epigenetic mechanisms.
Table 5.
Top molecular function annotations for the 21 genes nodes depicted in the nuclear compartment of the gene network shown in Figure 3.
| MOLECULAR FUNCTION ANNOTATION | P VALUE | # OF GENES |
|---|---|---|
| Transcription repressor activity | <1.0E-6 | 8 |
| Chromatin binding | 2.0E-6 | 6 |
| Protein deacetylase activity | 2.2E-2 | 2 |
| Histone deacetylase activity | 2.2E-2 | 2 |
| Deacetylase activity | 2.9E-2 | 2 |
DISCUSSION
A major and consistent observation of our previous genome-wide expression studies is that pediatric septic shock is characterized by early (i.e. within the first 24 hours of admission to the PICU) repression of genes corresponding to the adaptive immune system [1–4]. Assuming that that changes in gene expression have the potential to change cell function, this observation indicates that the pathobiology of pediatric septic shock may involve early dysfunction at the level of the adaptive immune system and is consistent, at the genomic level, with recent experimental and translational research paradigms [6, 14–21]. A potential confounding factor in these previous studies, however, is the use of whole blood-derived RNA leading to artificial repression of lymphocyte- and monocyte-specific genes secondary to reduced numbers of these leukocyte subsets in the context of septic shock. In other words, it is possible that our previous observations regarding suppression of adaptive immunity genes merely reflected reduced numbers of monocytes and lymphocytes, leading to a whole-blood trancriptome dominated by neutrophil-related gene expression patterns.
Herein we have directly addressed this important issue by conducting genome-wide expression studies using RNA derived from each of the three respective major leukocyte subsets, and comparing these data to the expression profiles from whole blood-derived RNA within the same patient cohort. The genes found to be differentially regulated in leukocyte subsets are enriched for metabolic and signaling pathways that are comparable to that found from whole-blood derived RNA. The data involving whole blood-derived RNA contain “signature probe sets” from all 3 major leukocyte subsets [13], and confirm early repression of genes corresponding to signaling pathways relevant to the adaptive immune system. More importantly, the data demonstrate that this gene repression profile remains evident when the expression studies are conducted using leukocyte subset-specific RNA. Thus, we have directly addressed one of the remaining major criticisms of our previous studies and can more confidently state that our previous observations represent valid gene expression profiles in pediatric septic shock, rather than epiphenomena of the experimental approach. Our data suggest that whole-blood derived RNA is a valid “tissue” on which to base genomic studies in patients with septic shock.
It was interesting to note that the gene expression profiles derived from lymphocyte-specific RNA contained the least number of differentially regulated genes compared to that of neutrophils and monocytes. This observation is unlikely to be a reflection of lymphopenia because the RNA amplification and labeling procedures were conducted using the same amount of RNA across all leukocyte subsets. Rather, this observation suggests that the lymphocyte population is relatively, and perhaps inappropriately, quiescent early in the course of pediatric septic shock. This assertion is well in line with current experimental paradigms of lymphocyte dysfunction in the context of septic shock [6, 14–21].
The gene network data also provide some interesting points for discussion and speculation in the context of recent literature. First, the top functional annotations associated with this gene network were directly related to processes involving the adaptive immune system. Since the network was derived via a merger of gene expression profiles across the three leukocyte subsets, the data suggest that adaptive immune dysfunction in the context of septic shock is a complex process involving all blood cell elements. Indeed, Freishtat and colleagues recently demonstrated that platelet-derived granzyme B plays a role in sepsis-associated lymphocyte apoptosis [22]. Second, it was interesting to note that IL-8 was one of the most highly connected gene nodes in the gene network. We recently demonstrated that serum IL-8 protein levels, measured within the first 24 hours of admission to the PICU, can be used to identify a population of children with septic shock having a very high likelihood of survival with standard care [5]. Third, CD163 was the most upregulated gene in the network. Although originally described as an important receptor for clearance of free hemoglobin from the circulation, very recent data indicate that CD163 also functions as a receptor for intact Gram-negative and Gram-positive bacteria [23]. Finally, it was interesting to note that the gene nodes localized to the nuclear compartment of the network corresponded to functional annotations related to transcriptional repression and chromatin modification, two processes that are directly related to epigenetic regulation of gene expression. The Kunkel laboratory has recently established the fascinating paradigm involving maladaptive epigenetic regulation of the immune system in the context of experimental sepsis [24].
The number of patients and controls in this study is relatively small relative to our previous studies. However, the primary objective of the current study was to validate our previous findings by specifically conducting genome-wide expression studies using leukocyte subset-specific RNA. Microarray data was complete across all patients and controls. Also, given that each study subject (control or patient) had 4 different types of RNA studies, the current study involves a total of 72 individual microarray chips.
Another potential limitation is whether or not this small cohort of patients is representative of the general population of pediatric patients with septic shock. A definitive statement is not possible, but the proportion of patients with a chronic disease (e.g. developmental delay, epilepsy, Prader-Willi syndrome) and the severity of illness (based on PRISM score) in the current cohort are similar to that of the largest interventional trial in pediatric septic shock to date [25].
In conclusion, we have validated genome-level repression of adaptive immunity gene programs early in the course of pediatric septic shock. These data further strengthen the evolving paradigm that human sepsis is not simply a problem of dysregulated innate immune function, but may also a problem of a dysfunctional adaptive immune system.
Supplementary Material
Acknowledgments
Supported by a grant from the National Institute of General Medical Sciences (RO1 GM064619).
REFERENCES
- 1.Wong HR, Cvijanovich N, Allen GL, Lin R, Anas N, Meyer K, Freishtat RJ, Monaco M, Odoms K, Sakthivel B, et al. Genomic expression profiling across the pediatric systemic inflammatory response syndrome, sepsis, and septic shock spectrum. Crit Care Med. 2009;37(5):1558–1566. doi: 10.1097/CCM.0b013e31819fcc08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong HR, Shanley TP, Sakthivel B, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Doctor A, Kalyanaraman M, Tofil NM, et al. Genome-level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol Genomics. 2007;30(2):146–155. doi: 10.1152/physiolgenomics.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shanley TP, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Doctor A, Kalyanaraman M, Tofil NM, Penfil S, Monaco M, et al. Genome-level longitudinal expression of signaling pathways and gene networks in pediatric septic shock. Mol Med. 2007;13(9–10):495–508. doi: 10.2119/2007-00065.Shanley. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cvijanovich N, Shanley TP, Lin R, Allen GL, Thomas NJ, Checchia P, Anas N, Freishtat RJ, Monaco M, Odoms K, et al. Validating the genomic signature of pediatric septic shock. Physiol Genomics. 2008;34(1):127–134. doi: 10.1152/physiolgenomics.00025.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong HR, Cvijanovich N, Wheeler DS, Bigham MT, Monaco M, Odoms K, Macias WL, Williams MD. Interleukin-8 as a stratification tool for interventional trials involving pediatric septic shock. Am J Respir Crit Care Med. 2008;178(3):276–282. doi: 10.1164/rccm.200801-131OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong HR. Pediatric septic shock treatment: new clues from genomic profiling. Pharmacogenomics. 2007;8(10):1287–1290. doi: 10.2217/14622416.8.10.1287. [DOI] [PubMed] [Google Scholar]
- 7.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437(7061):1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 8.Goldstein B, Giroir B, Randolph A. International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med. 2005;6(1):2–8. doi: 10.1097/01.PCC.0000149131.72248.E6. [DOI] [PubMed] [Google Scholar]
- 9.Pollack MM, Patel KM, Ruttimann UE. The Pediatric Risk of Mortality III--Acute Physiology Score (PRISM III-APS): a method of assessing physiologic instability for pediatric intensive care unit patients. J Pediatr. 1997;131(4):575–581. doi: 10.1016/s0022-3476(97)70065-9. [DOI] [PubMed] [Google Scholar]
- 10.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 11.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet. 2006;7(1):55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 12.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun L, Gorospe JR, Hoffman EP, Rao AK. Decreased platelet expression of myosin regulatory light chain polypeptide (MYL9) and other genes with platelet dysfunction and CBFA2/RUNX1 mutation: insights from platelet expression profiling. J Thromb Haemost. 2007;5(1):146–154. doi: 10.1111/j.1538-7836.2006.02271.x. [DOI] [PubMed] [Google Scholar]
- 14.Ayala A, Chaudry IH. Immune dysfunction in murine polymicrobial sepsis: mediators, macrophages, lymphocytes and apoptosis. Shock. 1996;6 Suppl 1:S27–S38. [PubMed] [Google Scholar]
- 15.Felmet KA, Hall MW, Clark RS, Jaffe R, Carcillo JA. Prolonged lymphopenia, lymphoid depletion, and hypoprolactinemia in children with nosocomial sepsis and multiple organ failure. J Immunol. 2005;174(6):3765–3772. doi: 10.4049/jimmunol.174.6.3765. [DOI] [PubMed] [Google Scholar]
- 16.Hotchkiss RS, Swanson PE, Freeman BD, Tinsley KW, Cobb JP, Matuschak GM, Buchman TG, Karl IE. Apoptotic cell death in patients with sepsis, shock, and multiple organ dysfunction. Crit Care Med. 1999;27(7):1230–1251. doi: 10.1097/00003246-199907000-00002. [DOI] [PubMed] [Google Scholar]
- 17.Hotchkiss RS, Tinsley KW, Karl IE. Role of apoptotic cell death in sepsis. Scand J Infect Dis. 2003;35(9):585–592. doi: 10.1080/00365540310015692. [DOI] [PubMed] [Google Scholar]
- 18.Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. N Engl J Med. 2003;348(2):138–150. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 19.Hotchkiss RS, Coopersmith CM, Karl IE. Prevention of lymphocyte apoptosis--a potential treatment of sepsis? Clin Infect Dis. 2005;41 Suppl 7:S465–S469. doi: 10.1086/431998. [DOI] [PubMed] [Google Scholar]
- 20.Hotchkiss RS, Osmon SB, Chang KC, Wagner TH, Coopersmith CM, Karl IE. Accelerated lymphocyte death in sepsis occurs by both the death receptor and mitochondrial pathways. J Immunol. 2005;174(8):5110–5118. doi: 10.4049/jimmunol.174.8.5110. [DOI] [PubMed] [Google Scholar]
- 21.Wheeler DS, Zingarelli B, Wheeler WJ, Wong HR. Novel pharmacologic approaches to the management of sepsis: targeting the host inflammatory response. Recent Pat Inflamm Allergy Drug Discov. 2009;3(2):96–112. doi: 10.2174/187221309788489779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freishtat RJ, Natale J, Benton AS, Cohen J, Sharron M, Wiles AA, Ngor WM, Mojgani B, Bradbury M, Degnan A, et al. Sepsis alters the megakaryocyte-platelet transcriptional axis resulting in granzyme B-mediated lymphotoxicity. Am J Respir Crit Care Med. 2009;179(6):467–473. doi: 10.1164/rccm.200807-1085OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fabriek BO, van Bruggen R, Deng DM, Ligtenberg AJ, Nazmi K, Schornagel K, Vloet RP, Dijkstra CD, van den Berg TK. The macrophage scavenger receptor CD163 functions as an innate immune sensor for bacteria. Blood. 2009;113(4):887–892. doi: 10.1182/blood-2008-07-167064. [DOI] [PubMed] [Google Scholar]
- 24.Wen H, Schaller MA, Dou Y, Hogaboam CM, Kunkel SL. Dendritic cells at the interface of innate and acquired immunity: the role for epigenetic changes. J Leukoc Biol. 2008;83(3):439–446. doi: 10.1189/jlb.0607357. [DOI] [PubMed] [Google Scholar]
- 25.Nadel S, Goldstein B, Williams MD, Dalton H, Peters M, Macias WL, Abd-Allah SA, Levy H, Angle R, Wang D, et al. Drotrecogin alfa (activated) in children with severe sepsis: a multicentre phase III randomised controlled trial. Lancet. 2007;369(9564):836–843. doi: 10.1016/S0140-6736(07)60411-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.