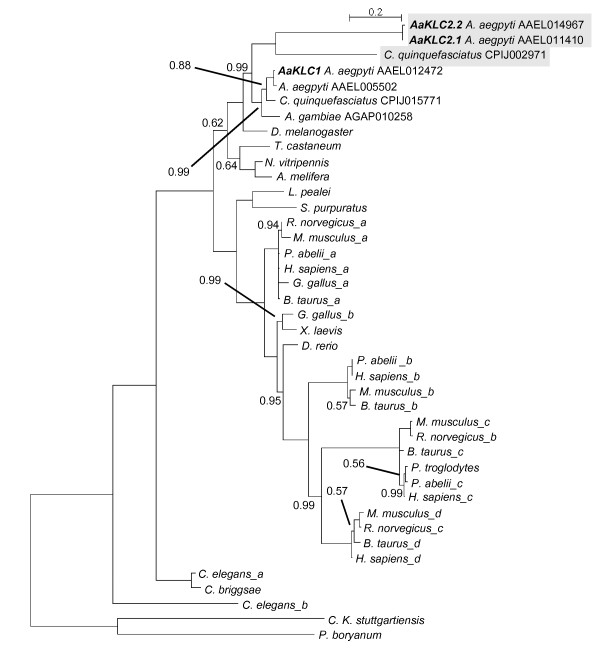
Figure 2.
Kinesin light chain phylogeny. Phylogenetic inference performed using MrBayes v3.1.2 with aa sequences conceptually translated from kinesin light chain genes of mosquitoes, other holometabolous insects, and divergent taxa. Clade credibility values are shown at each node only when they are not equal to 1.0. Accession numbers are given for mosquito sequences. Letters after species names indicate multiple KLC sequences for a species and are not associated with KLC nomenclature. See Additional File 3 for sequences and accession numbers, and Additional File 4 for alignment in FASTA format. Tree is rooted using kinesin light chain sequences from two bacterial species. Shaded sequences designate zygotic (AaKLC2.1 and AaKLC2.2) and potential zygotic (C. quinquefasciatus CPIJ00CP2971) genes. Bold letters highlight Ae. aegypti KLC genes. Scale shows number of aa changes per site. An. stephensi KLC1 is not included because we did not have the genome sequence at the time of phylogeny construction. AAEL005502 is not labeled AaKLC1.2 because this gene is a partial duplication of AaKLC1 and has only the Rab5-binding domain (see text).