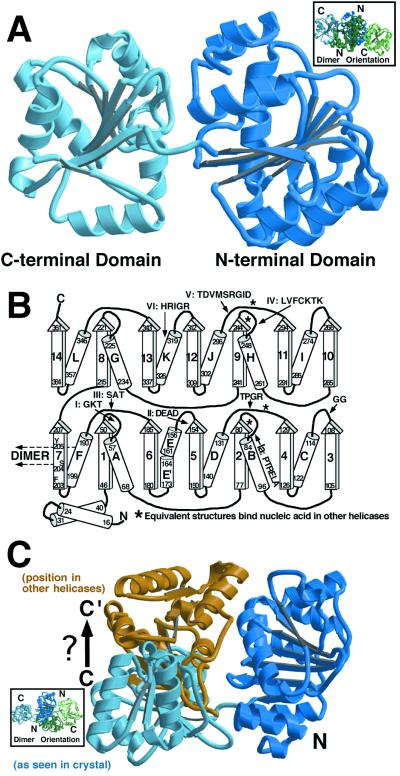
Figure 1.
Monomer structure. (A) The MjDEAD monomer showing the amino- and carboxyl-terminal domains (labeled N and C in subsequent figures). The linker between the domains can be seen in the middle of the figure. The orientation of the dimer in this view (in this and subsequent figures) is depicted in an Inset, with the equivalent domains colored blue, as in the main figure. Fig. 1A, as well as Fig. 1C, Fig. 2 A and B, and Fig. 3 A and C were made with MOLSCRIPT (41) and RASTER 3D (42). (B) The topological organization of the MjDEAD monomer, illustrating the similarities of the two domains. The “RecA-like core” stretches from β-strands 1 and 2 and 4–7 as numbered for the amino-terminal domain and their connecting α-helices. Sequence numbers at the edges of secondary structure elements are indicated, as are those loop regions observed to bind the nucleic acid backbone in some or all of known helicase complexes with nucleic acid. The region of polypeptide equivalent to the GG motif (motif 1B) also contacts nucleic acid in the HCV NS3 helicase. α-Helix F and β-strand no. 7 (that pack against their symmetry-related counterparts to form a dimer) are indicated. (C) Difference in the amino- and carboxyl-terminal domain orientation relative to other proteins. Superposition of only the amino-terminal domain with that of other proteins reveals a structure “opened up” relative to the others (blue domains). Independent superposition of the carboxyl-terminal domain on a “closed” structure (in this case the PcrA DNA and AMPPNP structure) leads to a closing of the MjDEAD structure to a conformation more like that observed for other helicases (blue amino-terminal domain and gold carboxyl-terminal domain). Single-stranded (ss)DNA binds at the top of the two domains in other helicases in this orientation. We assume that this closed structure for MjDEAD will likewise resemble the structure of the DNA/ATP bound form of this enzyme.