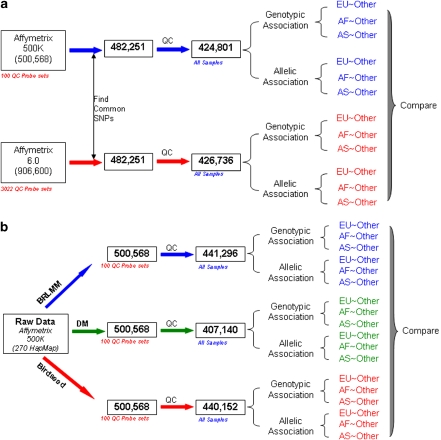
Figure 2.
Overview of the procedures for evaluating consistency between SNP arrays (a). Both data sets of the 270 HapMap samples from Affy500K and Affy6 were genotype called using algorithm Birdseed. The 482 251 SNPs interrogated in both arrays were used in the downstream association analysis. The same QC process was applied to both sets of genotypes before the same statistical tests for associations (see Materials and methods). Overview of the procedures for evaluating consistency between genotype calling algorithms (b). The raw data (CEL files) of Affy500K of the 270 HapMap samples were genotype called using algorithms DM, BRLMM, and Birdseed. The same QC process and the same statistical tests were used for the three sets of genotypes. In the association analysis, both allelic and genotypic association tests were conducted. Three different case–control frameworks were used: each of the three population groups (European, African, and Asia) was set as ‘case' with the other two as ‘control' (see Materials and methods).