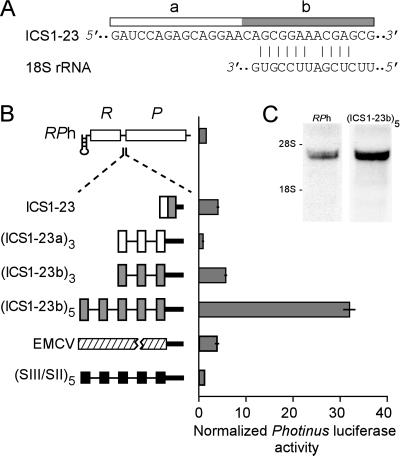
Figure 3.
Sequence and dicistronic analysis of selected sequence ICS1–23. (A) Complementarity between a segment of ICS1–23 and 18S rRNA. The upper sequence is that of ICS1–23, and the lower sequence is that of the rat 18S rRNA at nucleotides 1311–1324; complementary nucleotides are indicated by vertical lines. The open and gray bars indicate the nucleotide sequences that comprise sequences designated as ICS1–23a and ICS1–23b, respectively. (B) Dicistronic analysis of ICS1–23 and of multiple copies of ICS1–23a and ICS1–23b. A schematic representation of the dual luciferase construct used in this analysis is shown. Insets include one copy of selected sequence ICS1–23, three copies of ICS1–23a (open bars), and three or five copies of ICS1–23b (gray bars). Multiple copies of ICS1–23a and ICS1–23b are spaced nine nucleotides apart with poly(A) (thin lines) and separated from the Photinus initiation codon by 25 nt of the mouse β-globin 5′ untranslated region (thick black line). Other Insets include the EMCV IRES (hatched bar) and the (SIII/SII)β5 spacer control (12). This spacer control consists of five copies of a 9-nt segment from the mouse β-globin 5′ untranslated region (black bars) spaced 9 nt apart with poly(A). The histogram shows normalized Photinus luciferase activities (± SEM) (see Materials and Methods) in transfected B104 neuroblastoma cells. (C) Northern blot of total RNA purified from B104 cells transfected with either the RPh or (ICS1–23b)5/RPh dicistronic construct and probed with a Photinus luciferase riboprobe. The positions of the 28S and 18S rRNAs are indicated.