Figure 1.
Overview of the Methodology for TMD Analysis
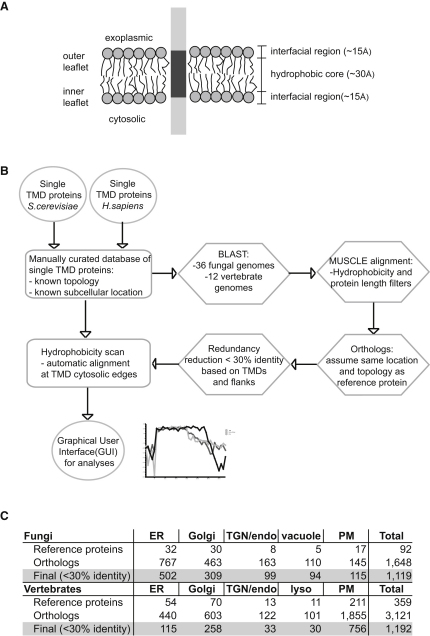
(A) Schematic of a typical single-pass or bitopic protein embedded in a lipid bilayer.
(B) Bitopic proteins of known topology and location from S. cerevisiae and H. sapiens were identified by literature and database searches. Orthologous proteins were identified using BLAST and aligned with the reference proteins. The starts of the TMDs were identified by a hydrophobicity scanning algorithm and used to align the TMDs at their cytosolic edges.
(C) The number of proteins from the indicated organelles that were used in the analyses of TMDs (PM, plasma membrane). Redundancy reduction was such that TMDs and flanking sequences have <30% identity. Reference proteins are listed in Table S1 and Table S2.
See also Figure S1.