Figure S2.
Analysis of the Significance and Robustness of Organelle-Specific Differences in TMD Hydropathy Plots, Related to Figure 3
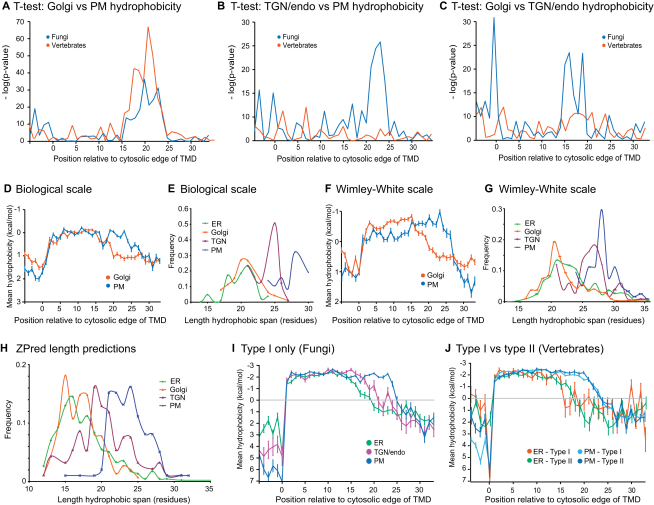
(A, B, and C) Independent (two sample) t tests were used to compare the mean residue hydrophobicity of residues at positions along the TMDs of proteins from (A) the Golgi and plasma membrane (PM); (B) TGN/endosomes and plasma membrane (PM); and (C) the Golgi and TGN/endosomes.
(D) Positional analysis of mean hydrophobicity of the Golgi and plasma membrane TMDs calculated using the Biological scale reported by Hessa and coworkers (Hessa et al., 2007). The Biological scale was also used to define cytosolic TMD edges and thus align the fungal TMDs from different organelles at their cytosolic ends. Error bars indicate standard error of the mean.
(E) Distribution of apparent lengths of fungal TMDs calculated by using the Biological scale to also define both the cytosolic and exoplasmic TMD edges.
(F and G) As for (D) and (E) except that the Wimley-White hydrophobicity scale was used to define the TMD edges and calculate the mean hydropathy at each position (Wimley and White, 1996).
(H) Distribution of lengths of TMDs obtained from the output of the TMD prediction program Zpred2 (Papaloukas et al., 2008). TMDs with 10 flanking residues on either side were used as the input for Zpred2 and the output was parsed to give a TMD length for each protein. In all cases a similar trend is seen: ER and Golgi TMDs are generally shorter than plasma membrane TMDs, with the TGN/endosome set being intermediate.
(I and J) Analysis of mean TMD hydropathy of proteins of different topology using the GES scale. Results are shown for type I fungal proteins from the ER, TGN/endosomes and plasma membrane (PM) datasets (I), and for vertebrate type I and type II proteins from the ER and plasma membrane datasets (J). Error bars indicate standard error of the mean.